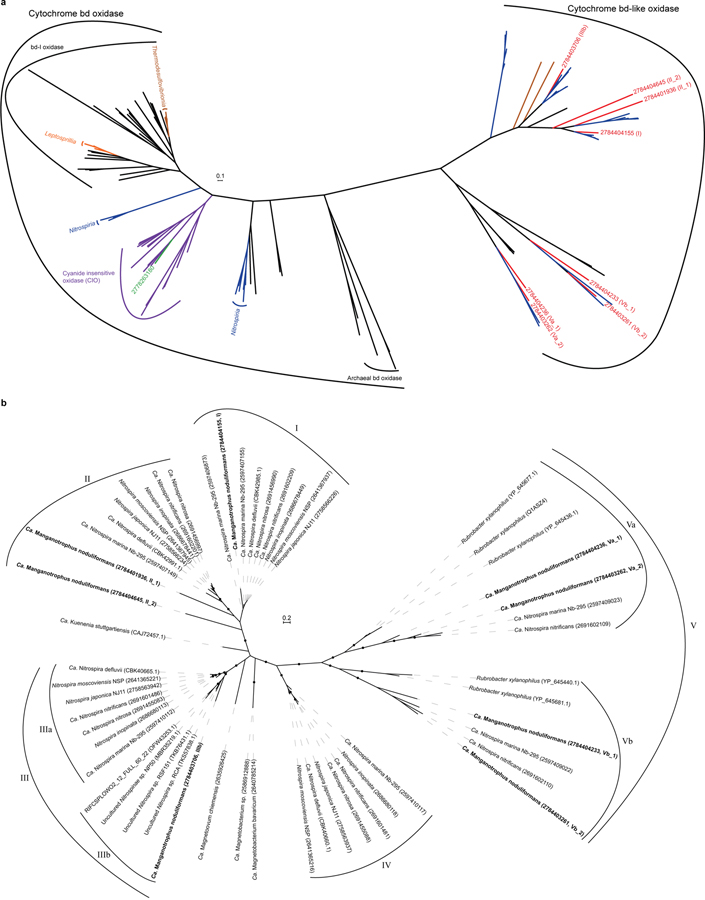
Extended Data Fig. 7 |. Phylogenetic analyses of cytochrome bd oxidase subunit I and cytochrome bd-like oxidases.
Only cytochrome bd-like oxidases were identified in Species A, in contrast to other classes in the phylum Nitrospirae (Nitrospirota). a, Unrooted maximum-likelihood tree, constructed using 242 amino acid positions shared between cytochrome bd and bd-like oxidases, using RAxML89 (model LGF). Deduced proteins from the genome of Species A are in red, with their IMG gene IDs and clade numbering (as shown in Fig. 3b) included in brackets. Other proteins from the phylum Nitrospirae (Nitrospirota) are coloured blue, orange or brown for classes Nitrospiria, Leptospirillia, or Thermodesulfovibrionia, respectively. Cytochrome bd oxidase of Species B, with its IMG ID, is in green; it belongs to the cyanide insensitive oxidase clade in purple. b, Phylogenetic analysis of cytochrome bd-like oxidases from Species A. Unrooted maximum-likelihood tree was constructed using 242 amino acid positions shared between different clades of cytochrome bd-like oxidases. Cytochrome bd-like oxidases are assigned to different clades, based on the phylogeny and their gene cluster structures. Species A encodes 8 cytochrome bd-like oxidases (bold), representing clades I, II, IIIb, Va and Vb; clade numbering as shown in Fig. 3b are included in brackets after the IMG IDs. Black dots on branches represent bootstrap values greater than 90%. Scale bars = evolutionary distance (substitutions-per-site average).