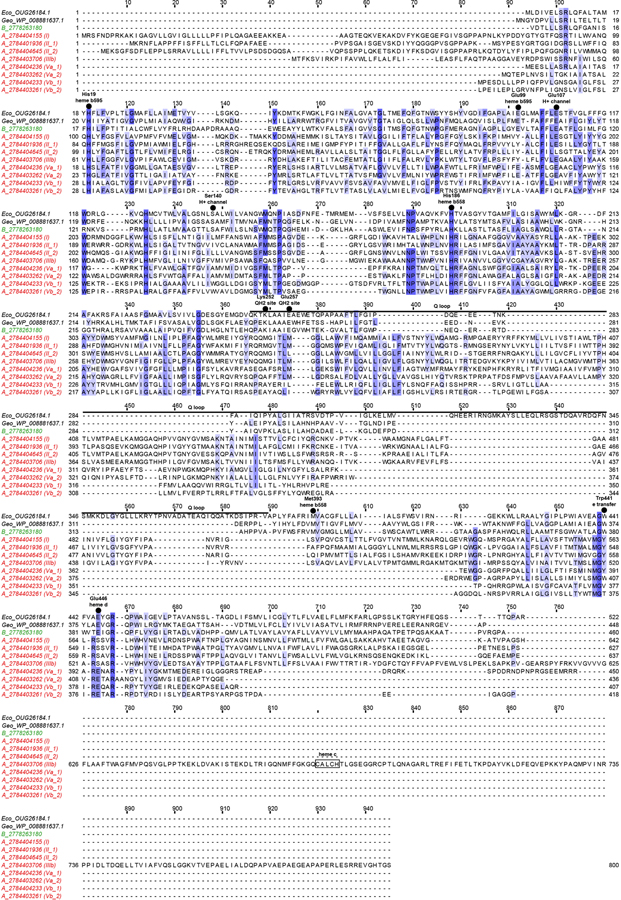
Extended Data Fig. 8 |. Sequence alignment of cytochrome bd and bd-like oxidases.
Cytochrome bd-like oxidase in Species A (sequence names starting with A, followed by their IMG gene ID and clade numbering as shown in brackets in Fig. 3b) and cytochrome bd oxidase subunit I in Species B (sequence name starting with B, followed by its IMG gene ID) are aligned to characterized cytochrome bd oxidases in Escherichia coli (sequence name starting with Eco, followed by its NCBI ID) and Geobacillus thermodenitrificans (sequence name starting with Geo, followed by its NCBI ID). Key features as revealed by structure102 are indicated at the top of the alignment, using E. coli protein residue numbering. The cytochrome bd oxidase subunit I sequence from Species B shows conservation of all key residues. In contrast, cytochrome bd-like oxidases in Species A do not show conservation of many key residues; instead, they are predicted to have 14 transmembrane helixes (compared to 9 in E. coli). One cytochrome bd-like oxidase in Species A has a C-terminus extension with a heme c binding motif (CXXCH).