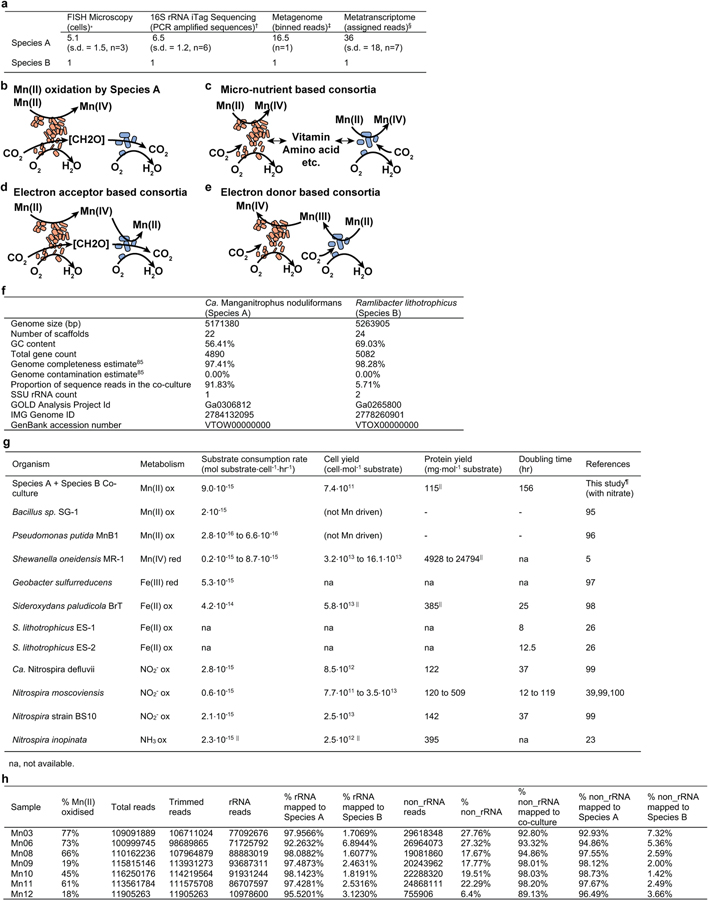
Extended Data Fig. 3 |. Properties of the refined co-culture.
a, Estimations of the relative ratio between Species A and Species B. *Slow-growing microbes, in particular Species A (which also has a smaller cell volume than Species B or Escherichia coli) could have lower number of ribosomes, resulting in lower signal intensity from rRNA-targeted fluorescent probes, relative to the fluorescent signal from DNA stain DAPI. †The two species together account for 99.7% of assigned sequence reads (Supplementary Table 1). ‡The two species together account for 97.54% of the sequence reads in the metagenome (panel f). §The two species together account for 99.576% (s.d. = 0.005%, n=7) of the rRNA sequence reads and 100.1700% (s.d. = 0.0005%, n=7) of the non-rRNA sequence reads in the co-culture metatranscriptomes (panel h). b-e, Possible metabolic interactions that may be occurring between Species A (orange) and Species B (blue). f, Genome statistics for Species A and Species B. g, Observed rates and yields of Mn(II) oxidation by the co-culture, in comparison to the literature values reported for other physiologically or phylogenetically related lithotrophs or metal-active heterotrophs. ||Conversion estimate based on Escherichia coli biomass of 2.8⋅10−13 g dry cell weight per cell, of which 55% is protein27. ¶Co-culture values correspond to results from the single independent culture with nitrate as N source for which extensive data on both oxidation kinetics and growth (genome copies) were collected. h, Transcriptome statistics for 7 co-cultures sampled at different degrees of Mn(II) oxidation.