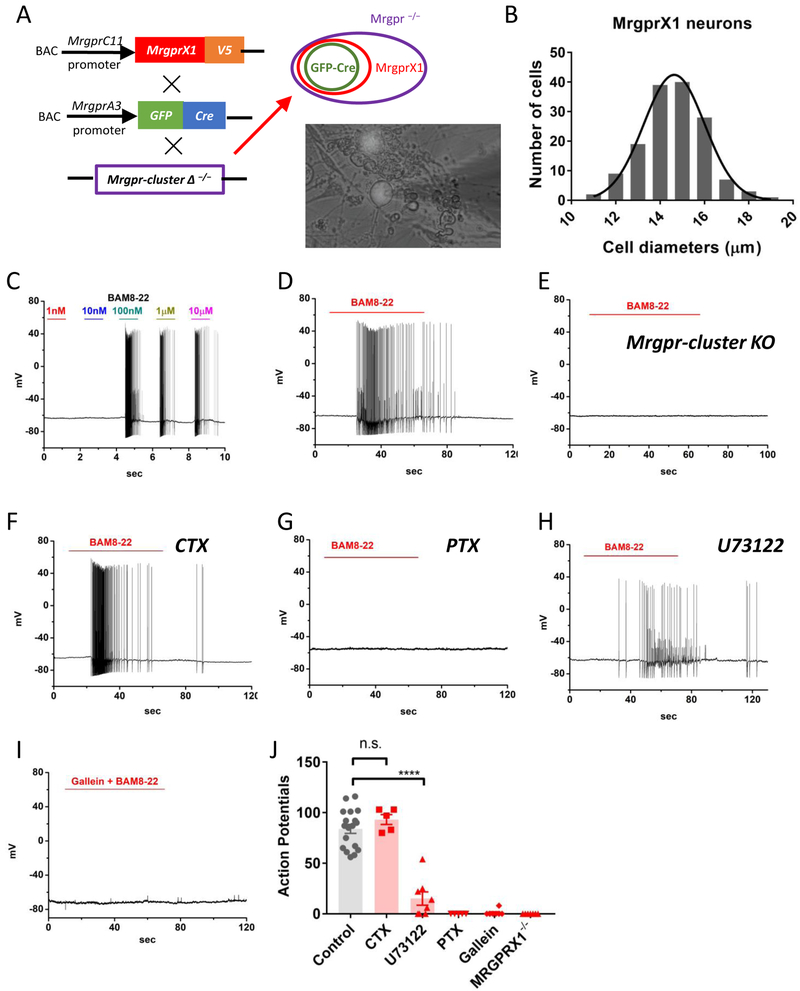
Figure 1. Activation of MrgprX1 triggers robust action potential discharges in dorsal root ganglion neurons.
(A) Strategy for generating the BAC transgenic line expressing human MrgprX1 driven by mouse MrgprC11promoter. The Mrgprc11 MrgprX1 transgenic mice were mated with Mrgpra3GFP-Cretransgenic mice and then crossed into the Mrgpr-cluster Δ−/−background. Since Mrgpra3 is a subpopulation of Mrgprc11, the GFP-cre will label a subset of MrgprX1 neurons. The purple circle indicates all DRG neurons that lacked the 12 endogenous mouse Mrgprs. The red circle indicates a subpopulation of all MrgprX1 neurons. The green circle indicates the neurons that expressed both MrgprX1 and Cre-GFP. The lower-right picture shows the MrgprX1 DRG neurons that expressed Cre-GFP in the nuclei. (B) The estimated sizes of MrgprX1 cells. The average diameter is 14.7 ± 0.11 μm (n=148). (C) A sample current-clamp recording of MrgprX1 neurons at different concentrations of BAM8-22 (n=3). (D) A sample recording of MrgprX1 neurons in the presence of 100nM for one minute (n=18). (E) A sample recording from Cre-GFP labeled, Mrgpr-cluster Δ −/−(MrgprX1−/−) DRG neurons in the presence of 100nM BAM8-22 (n=7). (F) Pretreatment with cholera toxin (2μg/mL) overnight had no effects on BAM8-22 evoked action potentials (n=5). (G) Pertussis toxin pretreatment (2μg/mL, overnight) completely block BAM8-22 evoked excitability (n=5). (H) Pretreatment with U73122 for 5 minutes greatly reduced, but did not prevent, BAM8-22 mediated excitability (n=8). (I) A Gβγ subunit inhibitor, Gallein (100μM), completely blocked BAM8-22 induced action potential discharges (n=8). (J) Summarized data for total action potentials discharges induced by 100nM BAM8-22 under different conditions. Data were presented as mean ± s.e.m., **** p=0.0001, by two-tailed t test.