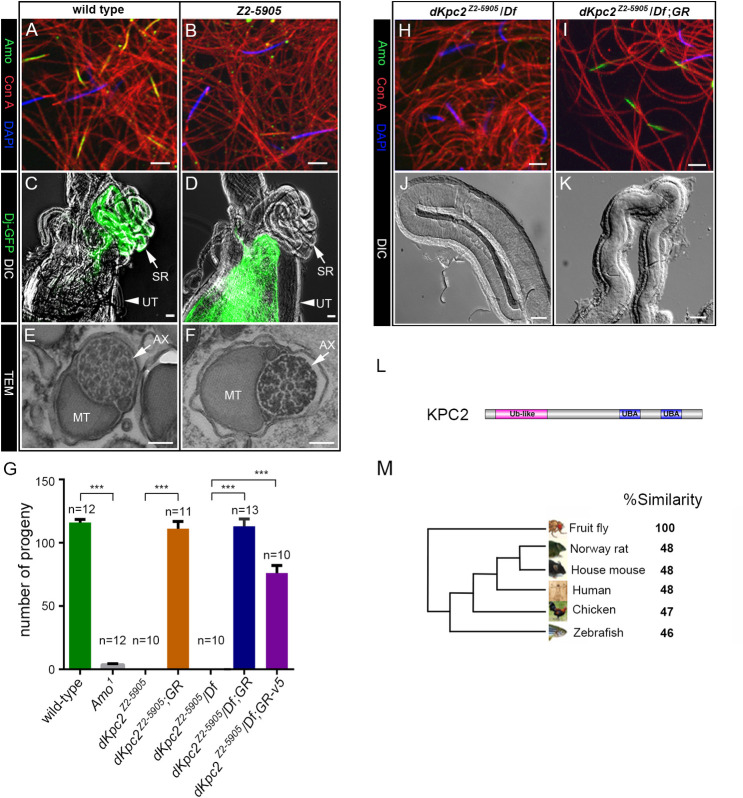
Fig 1. The Drosophila homolog of KPC2 is required for sperm tail localization of AMO.
(A-B) Sperm stained with anti-Amo: green, concanavalin A: red, DAPI: blue. Amo is missing from the sperm tail in Z2-5905 sperm (B). Scale bars: 5 μm. (C-D) Wild type female reproductive tracts dissected after mating with dj-GFP males (C) or Z2-5905; dj-GFP males (D). DIC and fluorescence images are super-imposed. Wild type sperm are found in the seminal receptacle (SR) while Z2-5905 mutant sperm are restricted to the uterus (UT). Scale bars: 100 μm. (E-F) Representative electron microscopy images from wild type (E) and Z2-5905 (F) sperm. Scale bars: 100 nm. MT: mitochondria AX: axoneme. (G) Fertility tests with males of the indicated genotypes mated to wild type females. The male fertility defect in Z2-5905 mutants is rescued by a CG11025 genomic rescue (GR) construct with or without a V5 tag. The number of tests per genotype is denoted above the bars. Df: deficiency. ***p<0.001. (H-I) Sperm stained with anti-Amo: green, concanavalin A: red, DAPI: blue. Scale bars: 5 μm. (J-K) Representative DIC images of wild type seminal receptacles dissected after mating with Z2-5905 males (J) or Z2-5905 males with a genomic rescue (K). The genomic construct rescues both Amo localization and sperm storage. Scale bars: 20 μm. (L) Predicted domain structure of all three KPC2 isoforms showing an N-terminal ubiquitin like domain (Ub-like) and two C-terminal ubiquitin associated domains (UBA). (M) Phylogenetic tree demonstrates homology between Drosophila KPC2 and related proteins.