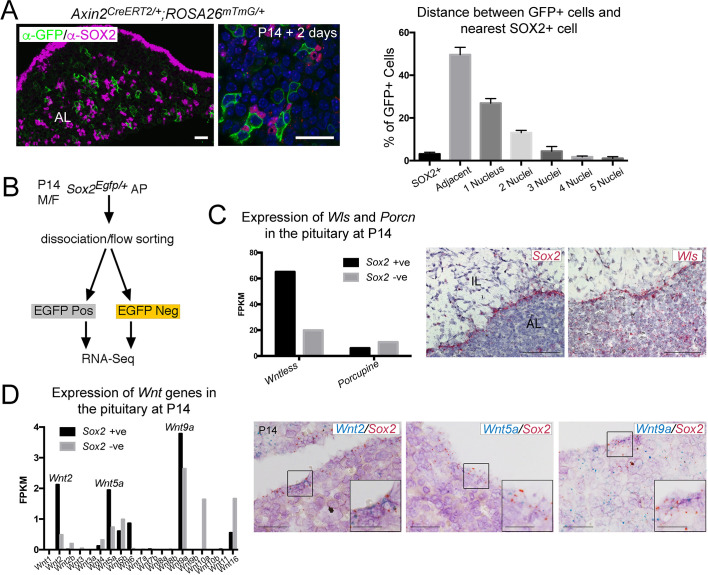
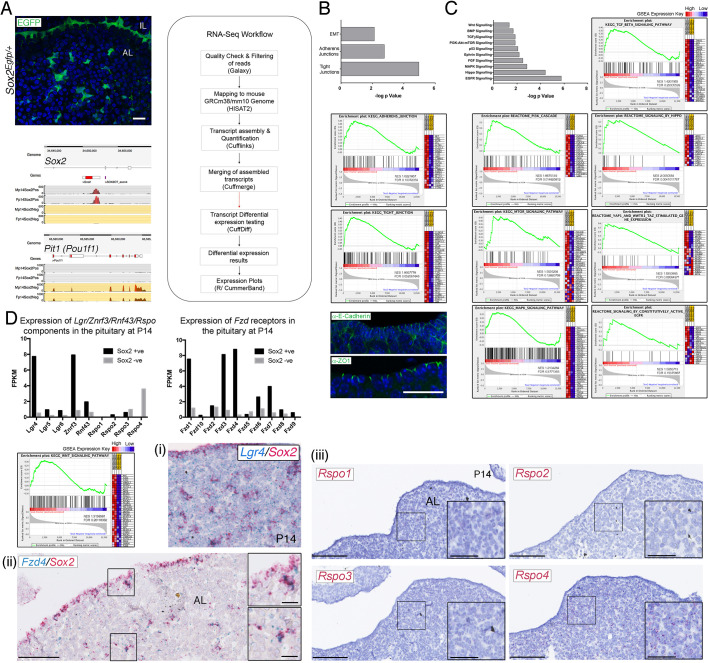
Figure 3. SOX2+ pituitary stem cells (PSCs) are as a source of WNT ligands in the pituitary.
(A) Immunofluorescence staining against GFP (green) and SOX2 (magenta) in Axin2CreERT2/+; ROSA26mTmG/+ pituitaries 48 hr post-induction. Graph representing a quantification of the proximity of individual GFP+ cells to the nearest SOX2+ cell as quantified by the number of nuclei separating them. Plotted data represents the proportion of GFP+ cells that fall into each category of the total GFP+ cells, taken from n = 3 separate pituitaries. Scale bars: 50 μm. (B) Experimental paradigm for RNA Seq analysis of Sox2 positive and negative cells. (C) Graphs representing the FPKM values of Wls and Porcupine in Sox2 positive and negative cells (black and grey bars, respectively). mRNA in situ hybridisation for Sox2 and for Wls on wild-type sagittal pituitaries at P14, demonstrating strong Wls expression in the marginal zone epithelium. Scale bars: 250 μm. (D) Bar chart showing the FPKM values of Wnt genes in the Sox2+ and Sox2− fractions. Double mRNA in situ hybridisation against Wnt2, Wnt5a, and Wnt9a (blue) together with Sox2 (red) validating expression in the Sox2+ population. Boxed regions through the marginal zone epithelium are magnified. Scale bars: 100 μm and 50 μm in boxed inserts.