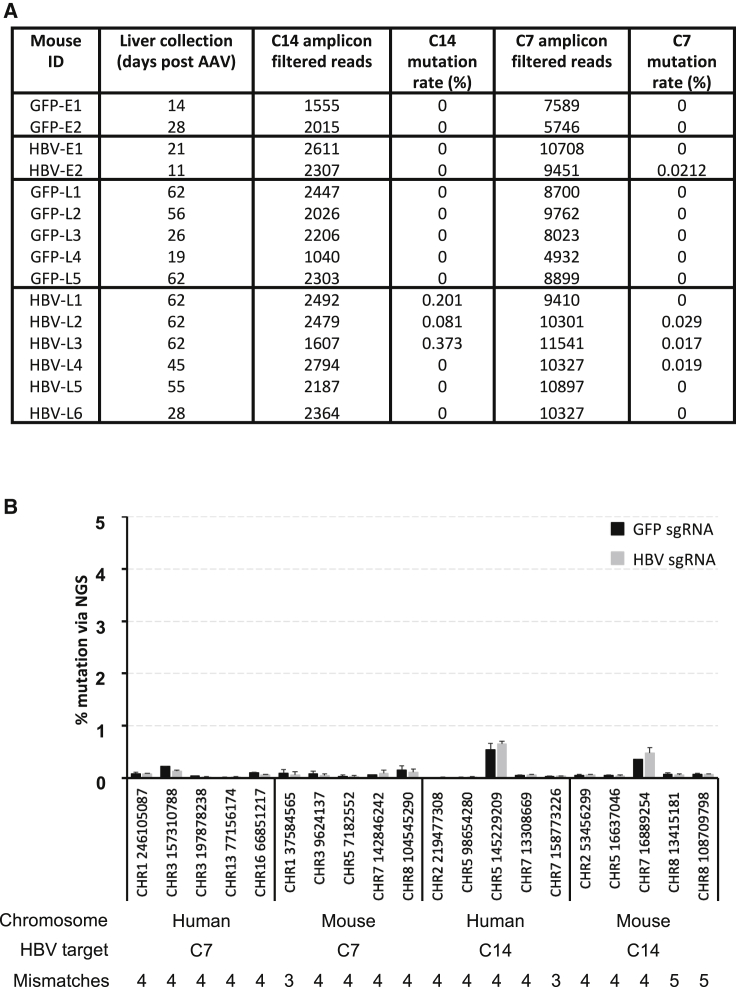
Figure 3.
HBV-specific and off-target gene editing
PCR amplicons spanning the C7 and C14 HBV target sites, or 20 closely related off-target sites for C7 or C14 within the human or mouse genomes (five of each) were amplified from liver genomic DNA and subjected to deep sequencing for analysis of mutations within the indicated 26-bp SaCas9 sgRNA target sequences. Mutations detected within each target site were identified using a custom script, and the mutation rate is based on mutations with multiple reads (non-singletons). (A and B) Mutation rates for the C7 and C14 target sites were determined for all mice (A), and the relative mutation rate for each off-target site was compared to the mutation rate of mice from the GFP-specific control sgRNA treatment group (B). The chromosomal location and starting nucleotide location for each off-target site are indicated along with the number of mismatches versus C17 or C14. Error bars indicate SD.