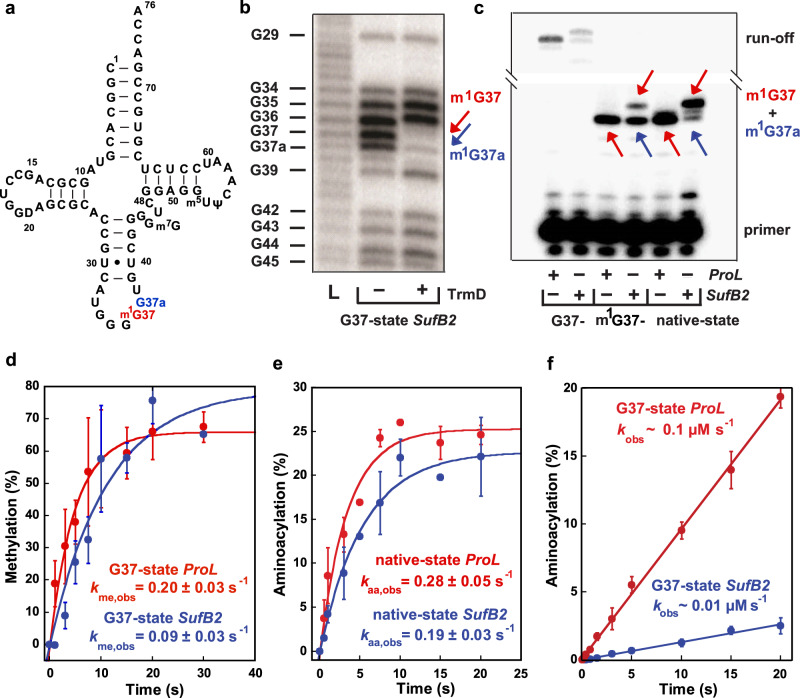
Fig. 1. Methylation and aminoacylation of SufB2 and ProL.
a Sequence and secondary structure of native-state SufB2, showing the N1-methylated G37 in red and the G37a insertion to ProL in blue. b RNase T1 cleavage inhibition assays of TrmD-methylated G37-state SufB2 transcript confirm the presence of m1G37 and m1G37a. Cleavage products are marked by the nucleotide positions of Gs. L: the molecular ladder of tRNA fragments generated from alkali hydrolysis. c Primer extension inhibition assays identify m1G37 in native-state SufB2. Red and blue arrows indicate positions of primer extension inhibition products at the methylated G37 and G37a, respectively, which are offset by one nucleotide relative to ProL. The first primer extension inhibition product for SufB2 corresponds to m1G37a, the second corresponds to m1G37, while the primer extension inhibition product for ProL corresponds to m1G37. Due to the propensity of primer extension to make multiple stops on a long transcript of tRNA, the read-through primer extension product (54–55 nucleotides) had a reduced intensity relative to the primer extension inhibition products (21–22 nucleotides). Molecular size markers are provided by the primer alone (17 nucleotides) and the run-off products (54–55 nucleotides). d TrmD-catalyzed N1 methylation of G37-state SufB2 and ProL as a function of time. e, f ProRS-catalyzed aminoacylation. e Aminoacylation of native-state SufB2 and ProL. f Aminoacylation of G37-state SufB2 and ProL as a function of time. In b, c, gels were performed three times with similar results, while in d–f, the bars are SD of three independent (n = 3) experiments, and the data are presented as mean values ± SD.