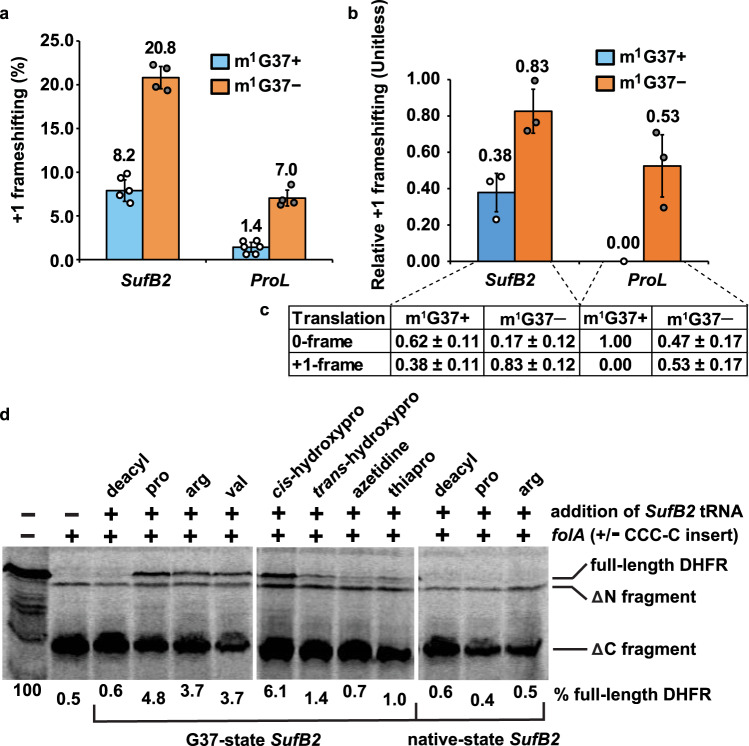
Fig. 2. SufB2-induced +1 frameshifting and genome recoding.
a The +1-frameshifting efficiency in cell-based lacZ assay for SufB2 and ProL strains in m1G37+ and m1G37– conditions. The bars in the graph are SD of 4, 5, or 6 independent (n = 4, 5, or 6) biological repeats, and the data are mean values ± SD. b The difference in the ratio of protein synthesis of lolB to cysS for SufB2 and ProL strains in m1G37+ and m1G37– conditions relative to ProL in the m1G37+ condition. c Measurements underlying the bar plots in (b). Each ratio was measured directly and the ratio of ProL in the m1G37+ condition was normalized to 1.0. The difference of each ratio relative to the normalized ratio represented the +1-frameshifting efficiency at the CCC-C motif at the 2nd codon of lolB. The bars in the graph are SD of 3 independent (n = 3) biological repeats, and the data are mean values ± SD. In a, b decoding of the CCC-C motif was mediated by SufB2 and ProM in the SufB2 strain, and by ProL and ProM in the ProL strain, where the presence of ProM ensured no vacancy at the CCC-C motif. The increased +1 frameshifting in the m1G37– condition vs. the m1G37+ condition indicates that SufB2 and ProL are each an active determinant in decoding the CCC-C motif. d SufB2-mediated insertion of non-proteinogenic amino acids at the CCC-C motif in the 5th codon position of folA using [35S]-Met-dependent in vitro translation. Reporters of folA are denoted by +/– CCC-C, where “+” and “–” indicate constructs with and without the CCC-C motif. SDS-PAGE analysis identifies full-length DHFR resulting from a + 1-frameshift event at the CCC-C motif by SufB2 pre-aminoacylated with the amino acid shown at the top of each lane, a ΔC fragment resulting from lack of the +1-frameshift event, and a ΔN fragment resulting from translation initiation at the AUG codon likely at position 17 or 21 downstream from the CCC-C motif. Gel samples were derived from the same experiment, which was performed five times with similar results. Gels for each experiment were processed in parallel. Lane 1: full-length DHFR as the molecular marker; deacyl: deacylated tRNA.