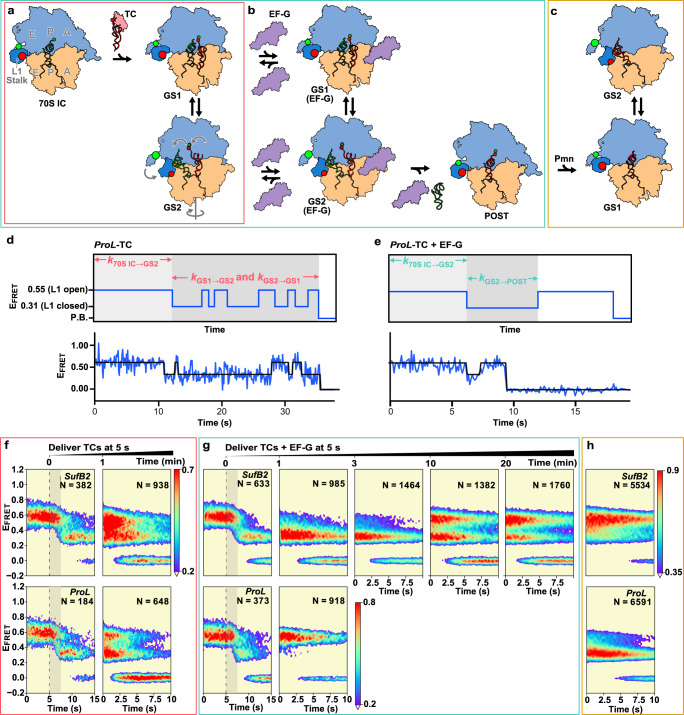
Fig. 6. SufB2 interferes with elongation complex dynamics during late steps of translocation.
a–c Cartoon representation of elongation as a G37-state SufB2- or ProL-TC is delivered to the A site of a bL9(Cy3)- and uL1(Cy5)-labeled 70 S IC; a in the absence, or b in the presence of EF-G, or c upon using puromycin (Pmn) to deacylate the P site-bound G37-state SufB2 or ProL and generate the corresponding PRE–A complex. The 30S and 50S subunits are tan and light blue, respectively; the L1 stalk is dark blue; Cy3 and Cy5 are bright green and red spheres, respectively; EF-Tu is pink; EF-G is purple; fMet-tRNAfMet is dark green; and SufB2 or ProL is dark red. d, e Hypothetical (top) and representative experimentally observed (bottom) EFRET vs. time trajectories recorded as ProL-TC is delivered to a 70S IC, d in the absence and e in the presence of EF-G as depicted in (a, b). The waiting times associated with k70S IC→GS2, kGS1→GS2, kGS2→GS1, and kGS2→POST are indicated in each hypothetical trajectory. f–h Surface contour plots of the time evolution of population FRET obtained by superimposing individual EFRET vs. time trajectories in the experiments in (a–c), respectively, for SufB2 (top) and ProL (bottom). N: the number of trajectories used to construct each contour plot. Surface contours are colored as denoted in the population color bars. For pre-steady-state experiments, the black dashed lines indicate the time at which the TC was delivered and the gray shaded areas denote the time required for the majority (54–68%) of the 70S ICs to transition to GS2. Note that the rate of deacylated SufB2 dissociation from the A site under our conditions is similar to that of EF-G-catalyzed translocation, thereby resulting in the buildup of a PRE complex sub-population over 3–20 min post-delivery that lacks an A site tRNA and is incapable of translocation. This sub-population exhibits kGS1→GS2, kGS2→GS1, and Keq values similar to those observed in experiments recorded in the absence of EF-G (Supplementary Table 6).