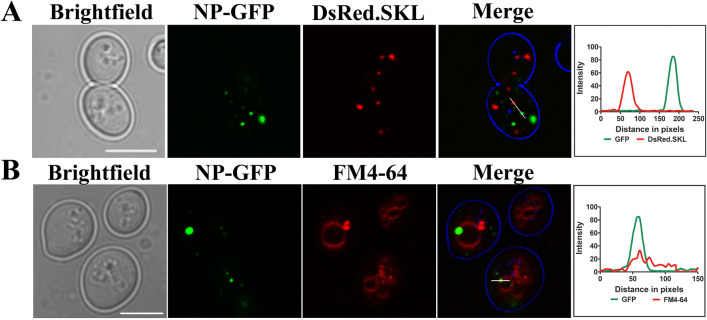
Fig. 3.
Subcellular localization of NP-GFP in yeast cells. (a) NP-GFP expressing cells co-transformed with DsRed-SKL plasmid to visualize peroxisomes. No colocalization, as also depicted by intensity profile generated by ImageJ is observed. NP-GFP expressing cells were incubated with FM4-64 (1 μg/μl), a vacuolar membrane staining dye for 1 h at 30 °C. Samples were subsequently analyzed by confocal microscopy. It is to be noted that not all NP-GFP puncta colocalize with the vacuolar marker. Lines drawn in merged panel are analyzed for the colocalization of GFP signal and DsRed.SKL or FM4-64 signal (red) in (a) and (b). Scale bar represents 5 µm. An overlap in the intensity profile obtained from the microscopy image is also depicted