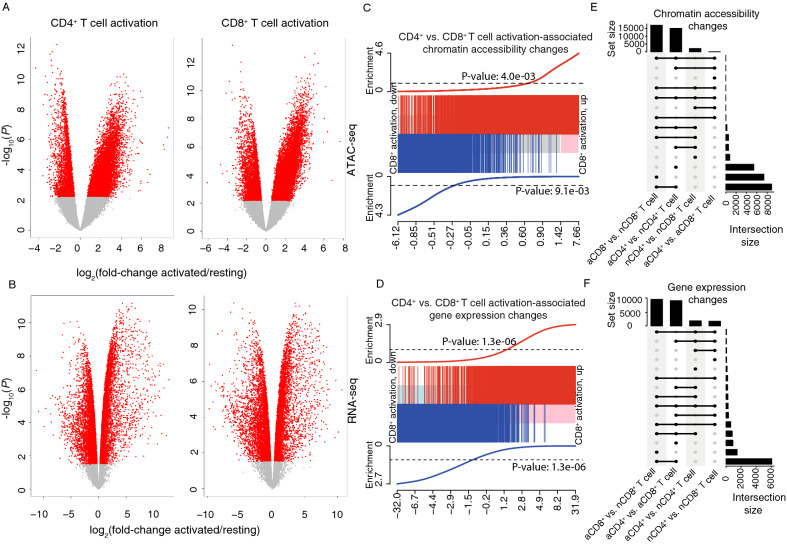
Figure 2.
T cell activation induces extensive changes in chromatin accessibility and gene expression that are comparable between CD4+ and CD8+ T cells. (A) Volcano plots showing the magnitude of differential accessibility in activated CD4+ and CD8+ T cells. Each point represents a peak with significant changes highlighted in red (FDR < 0.05). (B) Volcano plots showing the magnitude of differential expression in activated CD4+ and CD8+ T cells. Each point represents a gene with significant changes in red (FDR < 0.p05). (C) Barcode enrichment plot showing that DA peaks in CD4+ T cells are similarly ranked in CD8+ T cells. Peaks are ordered from left to right on the plot from most down-regulated to most up-regulated upon activation in CD8+ T cells, forming a shaded middle horizontal bar, with log2-fold-changes shown by the x-axis. DA peaks significantly up-or down-regulated upon activation in CD8+ T cells (FDR < 0.05) are marked by red and blue vertical bars, respectively. The red enrichment worm at the top of the plot shows the local density of red vertical bars while the blue enrichment worm at the bottom of the plot shows the local density for blue vertical bars. P-values show significance of the enrichments (fry gene set tests).
The enrichment pattern shows strong concordance of the CD4+ and CD8+ changes. (D) Barcode enrichment plot showing that DE genes in CD4+ T cells are similarly ranked in CD8+ T cells. (E) UpSet plot showing the number of DA peaks for each comparison (activated vs. resting CD8+ T cells, activated vs. resting CD4+ T cells, resting CD4+ vs. resting CD8+ T cells and activated CD4+ vs. activated CD8+ T cells) and their intersections. Vertical bars illustrate the total number of DA peaks for each comparison. Horizontal bars show the number of common DA peaks (intersection size) for a given set of comparisons (filled connected circles). (F) UpSet plot showing the number of DE genes for the same comparisons as in (E).