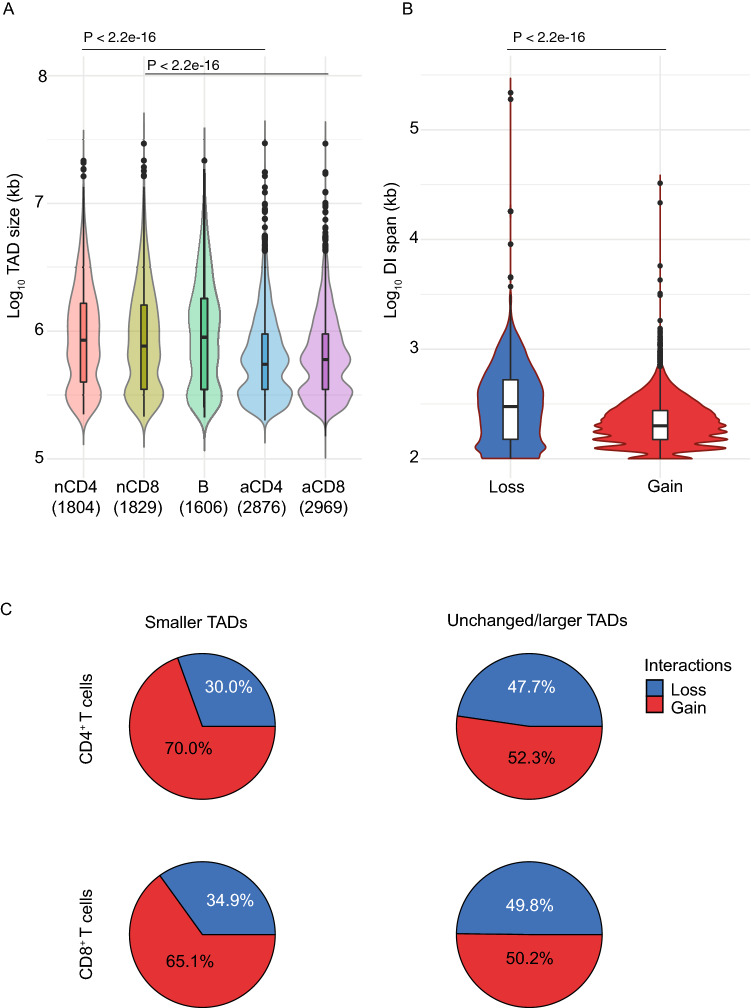
Figure 4.
T cell activation results in partitioning of genome topology. (A) Violin plots showing the distribution of TAD sizes in resting and activated T cells and B cells determined from summed libraries called with TADbit. Numbers in parenthesis depict the number of TADs called in each category. (B) Violin plots showing the distribution of the span of differential chromatin interactions (DIs) determined at 25 kbp resolution (FDR < 0.05) that are lost (decrease in logFC) or gained (increase in logFC) upon activation. Violin plots show median (horizontal line), interquartile range (boxes), values of the upper and lower quartiles (whiskers), outliers (circles) and kernel density estimation. Data for CD4+ and CD8+ T cells were merged as they showed a similar pattern. (C) Pie charts showing distribution of Loss and Gain chromatin interactions in TADs that become smaller in size (left) or remain unchanged/became larger in size (right) upon T cell activation. Statistical comparisons were made with the unpaired Wilcoxon test.