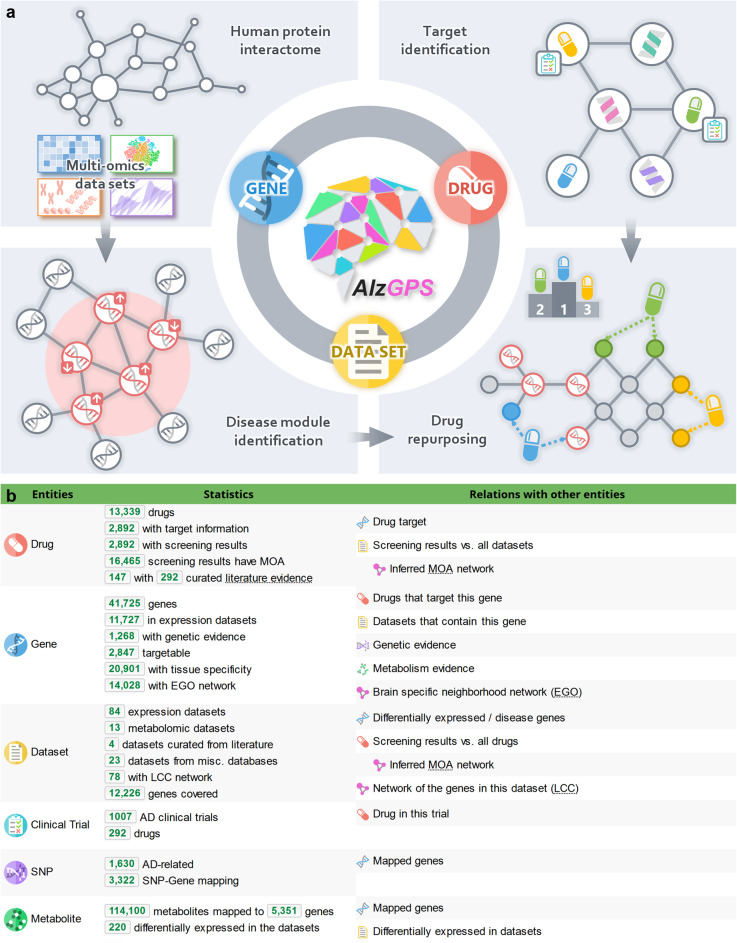
Fig. 1.
The architecture of AlzGPS. a AlzGPS was built on three main data entities (genes, drugs, and omics layers) and their relationships. The multi-omics data (genomics, transcriptomics (bulk and single cell/single nucleus), and proteomics) in AlzGPS help identify likely causal genes/targets that are associated with Alzheimer’s disease (AD) and disease modules in the context of human protein-protein interactome. Via network proximity measure between drug-target networks and disease modules in the human protein-protein interactome, drugs can be prioritized for their potential to alter the genes in the module for potential treatment of AD. b Detailed statistics of the entities and relations in AlzGPS. EGO, brain-specific neighborhood network (ego network); LCC, largest connected component network; MOA, mechanism-of-action network