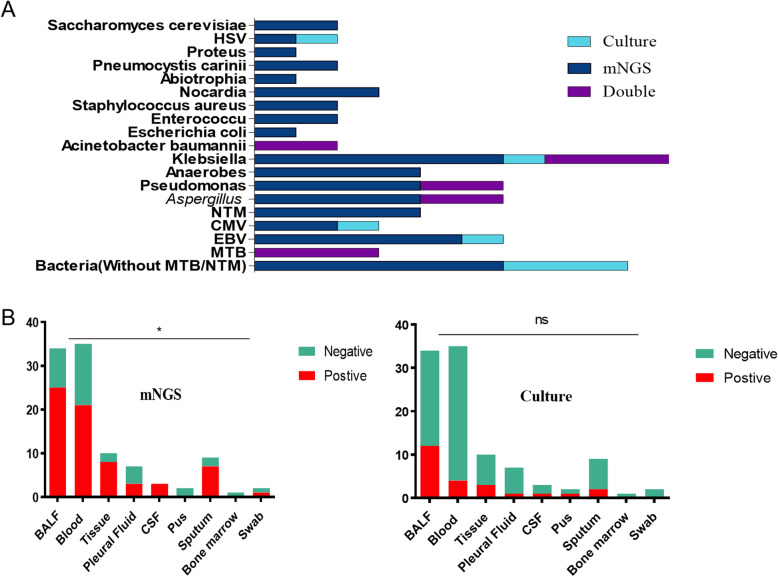
Fig. 5.
The overlap of positivity between mNGS and culture in pathogen and sample types. a. 19 pathogens detected in ID group with their corresponding frequencies were showed in histograms. Klebsiella, bacteria without MTB/NTM, EBV, CMV, NTM, Anaerobes, Saccharomyces cerevisiae, Proteus, Pneumocystis carinii, Abiotrophia, Nocardia, Staphylococcus aureus, Enterococcu and Escherichia coli demonstrated a trend of higher positivity rate in mNGS than that in culture with no statistical differences (P > 0.05). Acinetobacter baumannii and MTB were found equally in two groups. b. The overall sensitivity of mNGS in the different sample types were significantly different (P = 0.03) while sample types did not affect the sensitivity of pathogens in culture. Interestingly, especially in the types of BALF, blood and sputum samples, mNGS had significantly higher sensitivity than the culture (P = 0.002 for BALF, P < 0.001 for blood, P = 0.037 for sputum). Abbreviations: BALF, bronchoalveolar lavage fluid; CSF, cerebrospinal fluid; mNGS, metagenomic next-generation sequencing; HSV, herpes simplex virus; CMV, cytomegalovirus; EBV, Epstein-Barr virus; MTB, Mycobacterium tuberculosis; NTM, nontuberculous mycobacteria; ns, no significant difference