Fig. 1.
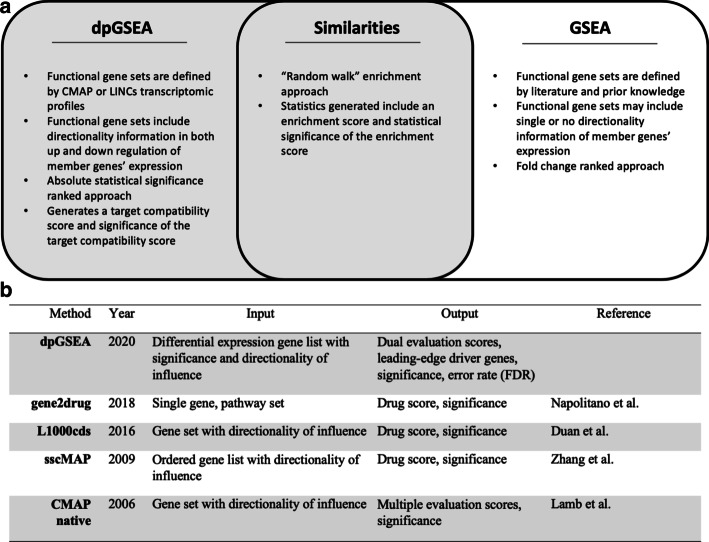
Comparisons between dpGSEA and similar approaches. a dpGSEA’s primary differences compared to GSEA include usage of a-priori gene set information derived from the Broad Institute’s connectivity map project (CMAP) and the library of integrated network-based cellular signatures (LINCS) projects organized as proto-matrices, an absolute statistical significance ranked approach rather than a fold change ranked approach, and a novel statistic to evaluate the drug target. Both approaches utilize a random walk running sum statistic to calculate enrichment scores. dpGSEA requires two inputs from the user to run. b dpGSEA is listed with comparable techniques that utilize GSEA-like approaches. Our approach uses the significance of a gene as well as directionality along with the generation of a novel statistic, the target compatibility score. We also show the driver genes for each drug