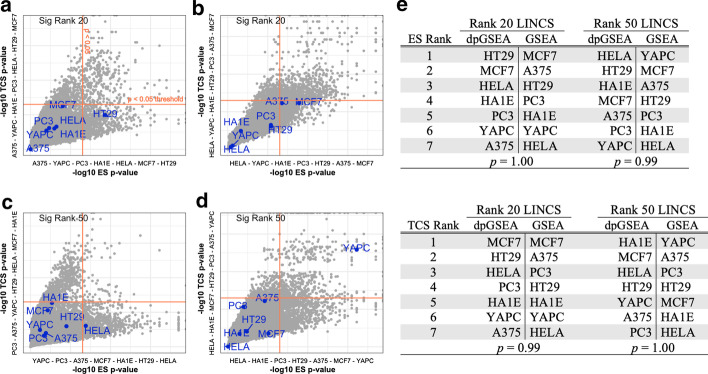
Fig. 3.
Comparisons between dpGSEA show statistically significant differences between enrichment results. Plots a–d show trends and comparisons between dpGSEA (a, c) and GSEA (b, d) for the top 20 and top 50 p value ranked proto-matrices (derived from LINCS data) identifying positively correlated genes. Plots a and b compare the top 20 ranked proto-matrices between dpGSEA and GSEA with each point representing an enriched drug in the final generated list. The labeled blue points all denote paclitaxel perturbated cell lines for the GEPNTs paclitaxel perturbation versus a GEPNTs DMSO control DE. The x-axis represents − log10 of the enrichment score (ES) p value and the y-axis − log10 of the target compatibility score (TCS) p value of corresponding perturbated drug cell line combination. The sub-axis lists the order of ascending significance for ES and TCS, respective of axis, that are also shown in tables E. The tables compare between the ranked orders for both ES and TCS with Wilcoxon signed rank test p values, suggesting the difference between dpGSEA and traditional GSEA results