Figure 3.
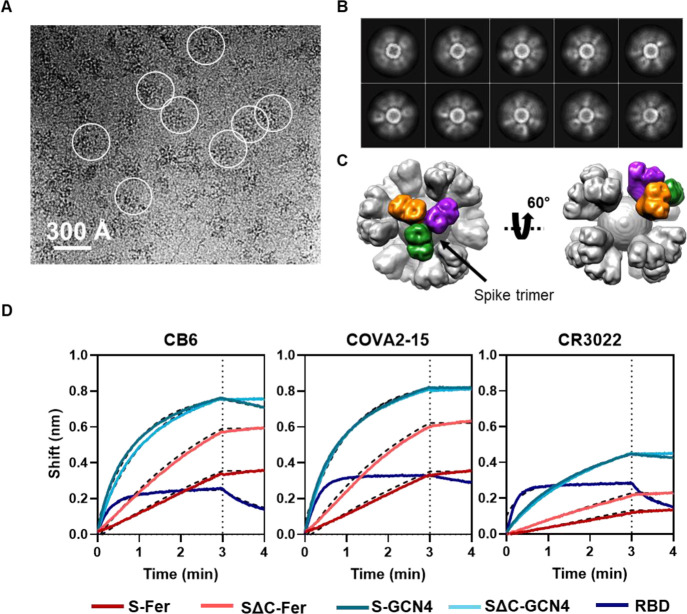
Cryo-EM and BLI confirm that spike proteins are presented on the particle surface with mAb epitopes intact. (A) Representative motion-corrected cryo-EM micrograph of the SΔC-Fer nanoparticles. Circles indicate representative particles that were picked for further analysis. Micrographs demonstrate that particles are approximately 300 Å. (B) Reference-free 2D class averages of SΔC-Fer. 2D class averages confirm the presence of both ferritin particles and the display of spike on the surface seen as density surrounding the particles. (C) Reconstructed cryo-EM map of the SΔC-Fer nanoparticle in two views. A single spike trimer on the surface is highlighted with each protomer of the trimer shown in a different color. (D) BLI binding of SARS-CoV-2 mAbs to purified spike antigens. Antigens were diluted to 100 nM monomer concentration (100 nM RBD, 33.3 nM S-GCN4 and SΔC-GCN4 trimer, and 4.2 nM S-Fer and SΔC-Fer 24-mer ferritin particle). Binding of all antigens to three SARS-CoV-2 reactive mAbs indicates that spike ferritin nanoparticles display epitopes similarly to the RBD and spike trimers. Curves were fitted with an association/dissociation nonlinear regression, and fits are represented with dashed black lines; kon values for each binding reaction are shown in Figure S4A. Both S-Fer and SΔC-Fer exhibited a slight increase in signal during the dissociation step, perhaps due to rearrangements of the particles on the BLI sensor tip due to the extensive avidity present on the multimerized particles. Lack of binding to an off-target Ebola-specific antibody (ADI-15731) is presented in Figure S4B. Binding experiments were performed in at least duplicate; a representative trace and fit are shown from one replicate.