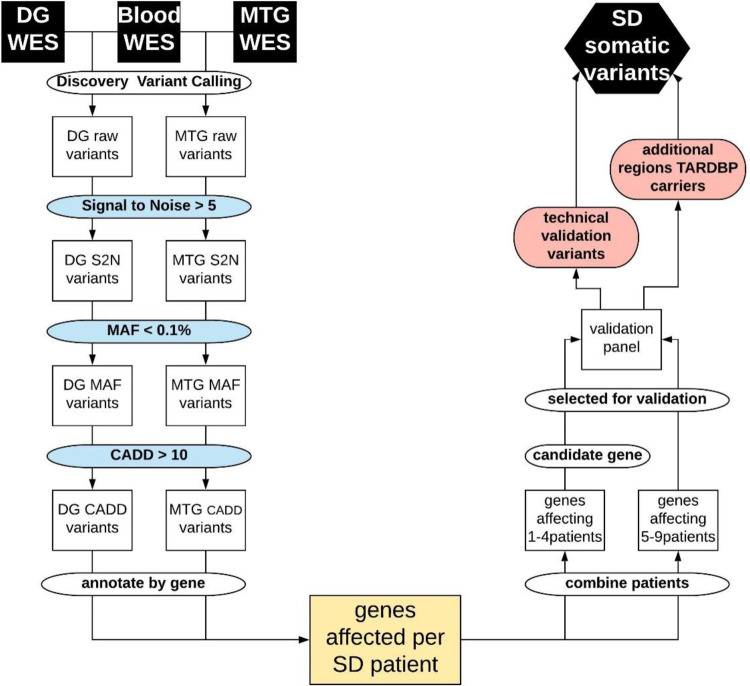
Figure 1.
Flowchart of data filtering and analysis. From top left: Raw somatic variant calling using blood and dentate gyrus (DG) or medial temporal gyrus (MTG) deep exome sequencing data (WES), signal to noise filter (S2N), minor allele frequency filter (MAF), CADD score filter, annotating and grouping per gene, resulting in the genes affected in each patient with semantic dementia (SD). Right: Grouping genes affecting multiple patients (>5) or affecting candidate genes in fewer patients (one to four) to be included in the validation amplicon panel. To excluded false negative findings in the WES data in FTD-TDP known germline causal genes GRN and TARDBP, all exons in these genes were included in the validation panel. The first validation round was performed on the same tissues as the discovery WES to confirm true positive variants from the WES, or identify false negative findings in GRN or TARDBP. The second round of validation further classified true positive variants in additional brain tissues and non-demented controls.