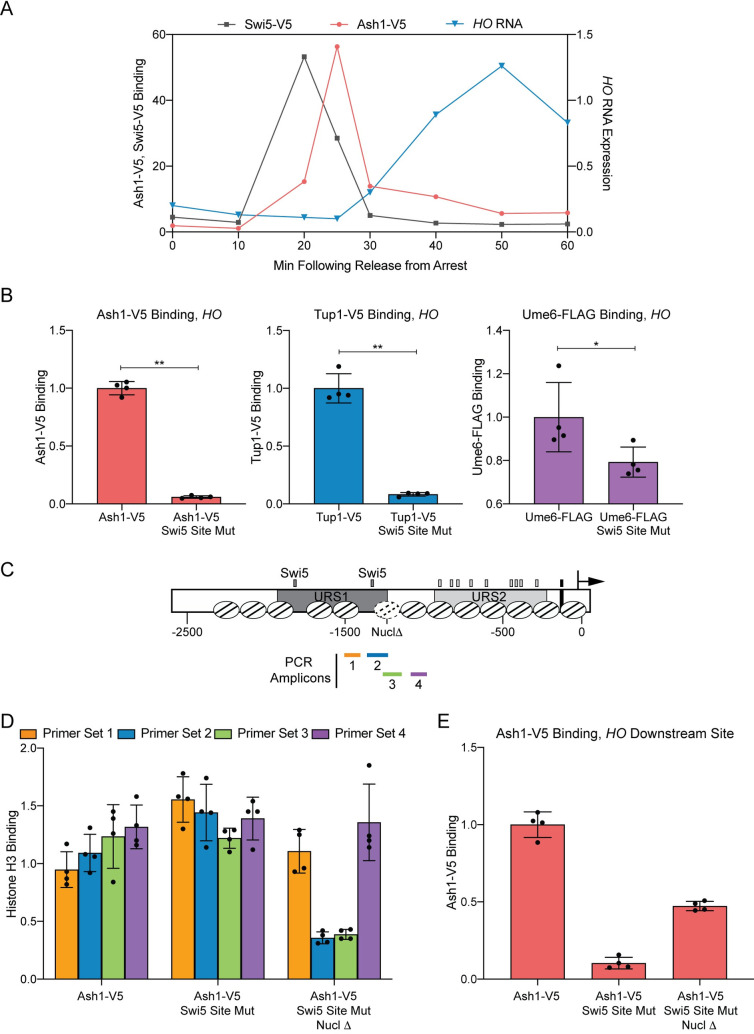
Fig 7. Binding of the Ash1 repressor to the HO promoter only occurs under conditions of low nucleosome density.
(A) Cell cycle time course of Swi5 binding, followed by Ash1 recruitment, and finally, HO expression. ChIP and HO mRNA analysis were performed in Swi5-V5 or Ash1-V5 strains containing the GALp::CDC20 allele and synchronized by galactose withdrawal and readdition. The 0 min time point represents the G2/M arrest, before release with galactose addition. Cells were harvested at the indicated time points following release (x-axis). Binding of Swi5 (gray; HO -1429 to -1158; left y-axis) and Ash1 (red; HO -1295 to -1121; left y-axis) was normalized to enrichment at an intergenic region on chromosome I (IGR-I) and to the corresponding input sample. HO mRNA levels (blue; right y-axis) were normalized to RPR1. (B) Swi5 binding is required for Ash1 binding and Tup1 recruitment. Ash1-V5, Tup1-V5 and Ume6-FLAG ChIP analysis, followed by qPCR with primers from HO -1295 to -1121. “Swi5 Site Mut” indicates strains in which both Swi5 binding sites are mutated and nonfunctional for HO activation. Binding at HO for each sample was normalized to its corresponding input DNA and to a positive reference control [CLN3 for Ash1, TEC1 for Tup1 and INO1 for Ume6; 27]. Each dot represents a single data point, and error bars reflect the standard deviation. ** p < 0.01, * p < 0.05. (C) Schematic of the HO promoter with positions of nucleosomes from MNase-Seq shown as ovals with slanted lines. The “Nucl Δ” nucleosome with dotted lines indicates the -1215 nucleosome targeted for displacement by introduction of two Reb1 sites (TTACCC) that substitute for HO sequences from -1268 to -1262 and from -1194 to -1189. Positions of the PCR amplicons are indicated with colored bars. (D) H3 ChIP shows Reb1 sites lead to nucleosome loss. Graph shows histone H3 ChIP analysis using strains that are Ash1-V5 with Swi5 wild type binding sites (Ash1-V5) or Swi5 binding site mutations (Ash1-V5 Swi5 Site Mut) or Swi5 binding site mutations and nucleosomal substitutions with Reb1 sites to displace the nucleosome (Ash1-V5 Swi5 Site Mut Nucl Δ). qPCR was performed with ChIP material using the following primers: primer set 1 (orange) = HO -1497 to -1399; primer set 2 (green) = HO -1347 to -1248; primer set 3 (blue) = HO -1257 to -1158; primer set 4 (purple) = HO -1277 to -978. Binding at each HO site was normalized to an intergenic region on chromosome I (IGR-I) and to the corresponding input DNA and the No Tag control. ** p < 0.01, * p < 0.05. (E) Nucleosome loss partially restores Ash1 binding even in the absence of the normally required Swi5 activator. Ash1 binding was measured by ChIP, using the same chromatin samples as the histone H3 ChIP in D. Binding in each sample was measured by qPCR at HO -1295 to -1121 and normalized to the CLN3 positive reference control and its corresponding input DNA. ** p < 0.01.