Figure 4. EEEV-33 recognizes a critical domain A epitope on SINV/EEEV particles for inhibition of viral entry or fusion.
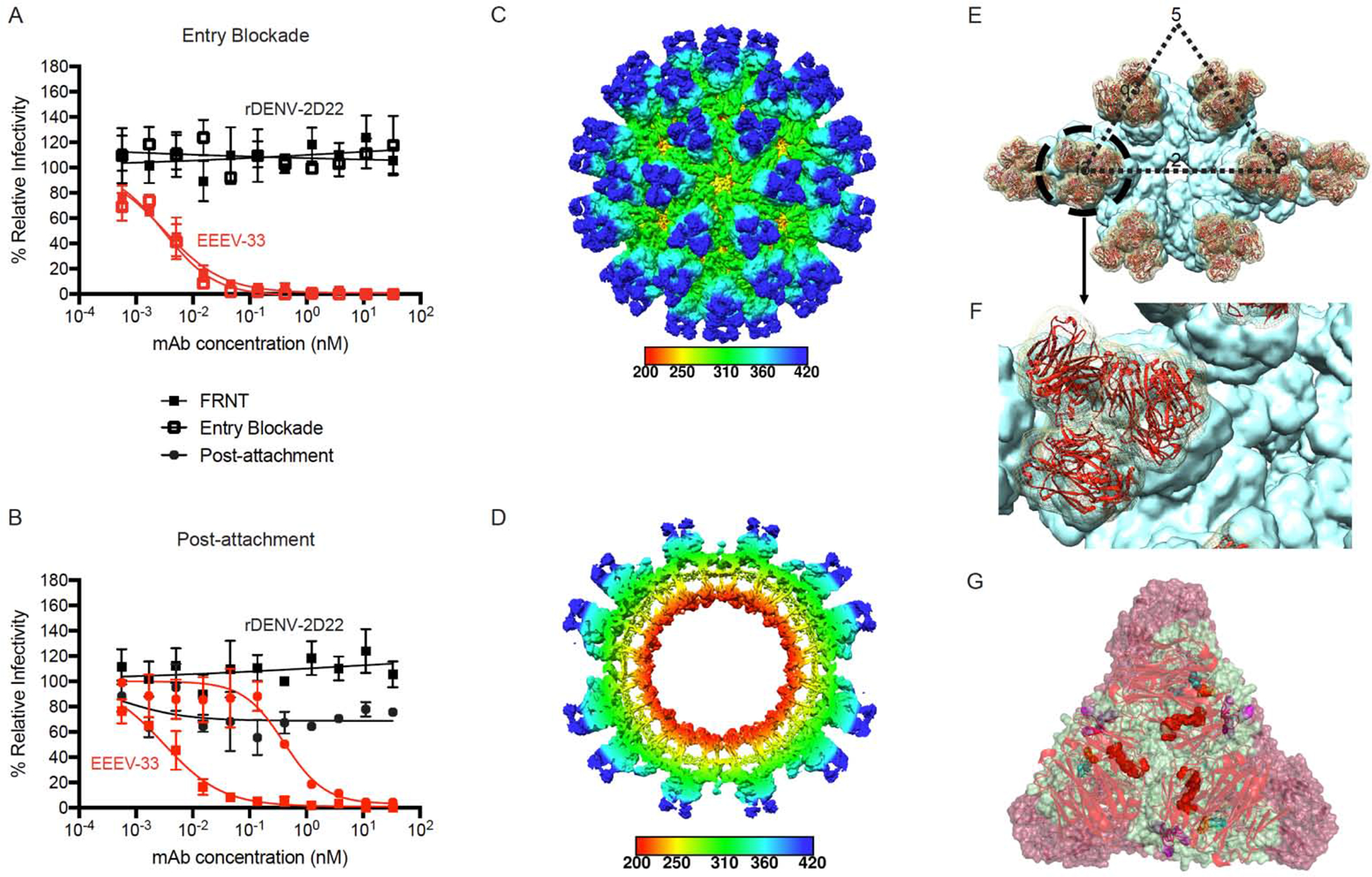
(A) Entry blockade of SINV/EEEV by EEEV-33. An entry blockade assay (open squares) was performed by extensive washing of EEEV-33 (red) or the control mAb (black), from the medium following internalization of SINV/EEEV into Vero cells. Representative neutralization curves are shown for EEEV-33 and rDENV-2D22 as determined through FRNT (closed squares; see Figure 1A) or the entry blockade assay with mAb concentration (nM) on the x-axis and percent relative infectivity on the y-axis.
(B) Post-attachment neutralization of SINV/EEEV. A post-attachment neutralization assay (starred circles) was performed by incubation of Vero cells with SINV/EEEV at 4°C for 1 hour followed by addition of EEEV-33 (red) or rDENV-2D22 (black) at 4°C for 1 hour. Cells were incubated at 37°C for 15 min prior to addition of an overlay and incubation at 37°C for 18 h. Representative neutralization curves are shown for EEEV-33 and rDENV-2D22 as determined through FRNT (closed squares; see Figure 1A) or the post-attachment neutralization assay with mAb concentration (nM) on the x-axis and percent relative infectivity on the y-axis.
(C–D) Cryo-EM reconstruction of SINV/EEEV in complex with EEEV-33 Fab. Cryo-EM structure of EEEV-33 Fab complex (~7.2 Å) showing radially colored surface representation of full (C) and cross section (D) of the map.
(E) EEEV-33 Fab binding footprint to E2 trimeric spikes on SINV/EEEV particles. View of map surface to illustrate binding of EEEV-33 Fab (red) to the q3 and i3 spikes along the icosahedral 2-fold axis.
(F) EEEV-33 Fab constant domain contact interactions. Close-up view of EEEV-33 Fab binding to the i3 spike (black circle in E), in which overlapping Fab constant domain density is observed.
(G) Cryo-EM E2 trimeric view of EEEV-33 Fab binding with critical alanine residues.
Critical alanine residues identified for EEEV-33 are indicated with spheres to illustrate the epitope of EEEV-33 corresponds with the SINV/EEEV (PDB ID: 6MX4) and EEEV-143 Fab (mutated sequence of PDB: 6MWX) docked and rigid body refined cryo-EM model of rEEEV-33 Fab in complex with SINV/EEEV. Sphere color corresponds to E2 domain (N-link - purple, Domain A - red, Arch 1 - magenta, Domain B - cyan, and Arch 2 - orange) as described in Figure 3D.
Data in A–B represent mean ± SD of technical triplicates and are representative of two independent experiments. See Figure S4 for additional views of EEEV-33 Fab binding to the E2 trimer on SINV/EEEV particles.
