Fig. 2. USP16 specifically interacts with IKKβ but not p105 or IκBα.
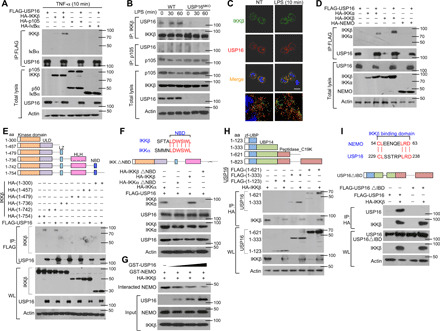
(A) HEK293T cells were transfected with USP16-, IKKβ-, p105-, and IκBα-expressing plasmids. IB of HA was performed followed by IP with an anti-FLAG antibody on WLs. (B) The interaction among USP16, IKKβ, and p105 was assessed in WT and USP16-deficient BMDMs activated by LPS. WLs were subjected to IP using an anti-IKKβ or anti-p105 antibody and then to IB analyses of the associated USP16. (C) Confocal microscopy analysis of the colocalization of USP16, IKKβ, and DAPI in WT macrophages stimulated with LPS (100 ng/ml) as indicated. Scale bar, 5 μm. NT, nontreatment. (D) HEK293T cells were transfected with the indicated plasmids. The interaction between USP16 and IKK components was evaluated by co-IP assay. (E and F) The associations between USP16 and various truncation mutants of IKKβ (E) or a USP16 interaction-defective mutant (IKKβ∆NBD) (F) were detected through the indicated IP and IB analyses. (G) USP16 competently bound to IKKβ and inhibited the interaction between IKKβ and NEMO. (H and I) The binding amounts of IKKβ and various truncation mutants of USP16 (H) or an IKKβ interaction-defective mutant (USP16∆IBD) (I) were detected by the indicated IP. The data are representative of at least three independent experiments. aa, amino acids.
