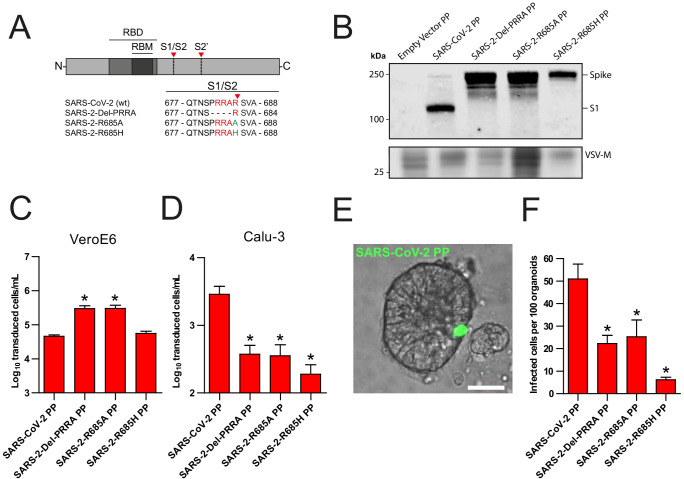
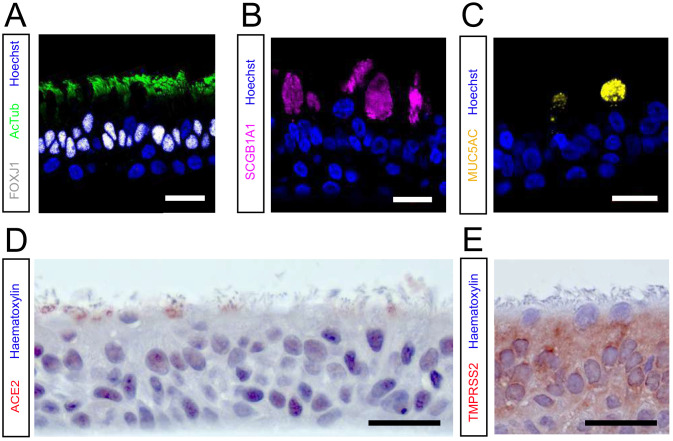
Figure 1. The SARS-CoV-2 S multibasic cleavage site mediates entry into human airway organoids.
(A) Schematic overview of SARS-CoV-2 S protein mutants. MBCS residues are indicated in red; amino acid substitutions are indicated in green. Red arrows indicate cleavage sites. The SARS-CoV-2 S MBCS was mutated to either remove the PRRA motif (SARS-2-Del-PRRA) or substitute the R685 site (SARS-2-R685A and R685H). (B) Comparison of S cleavage of SARS-CoV-2 PPs and the MBCS mutants. Western blots were performed against S1 with VSV-M silver stains as a production control. (C and D) PP infectivity of SARS-CoV-2 S and MBCS mutants on VeroE6 (C) and Calu-3 (D) cells. (E) Differentiated hAO cultures were infected with concentrated SARS-CoV-2 PPs containing a GFP reporter, indicated in green. Scale bar indicates 20 μm. (F) SARS-CoV-2 PP and MBCS mutant infectivity on bronchiolar hAO cultures. One-way ANOVA was performed for statistical analysis comparing all groups with SARS-CoV-2 PPs. Experiments were performed in triplicate (B and D, F). Representative experiments from at least two independent experiments are shown (C and D). Combined data from three independent experiments is shown (F). Error bars indicate SEM. *p<0.05. GFP, green fluorescent protein; hAO, human airway organoid; MBCS, multibasic cleavage site; PP, pseudoparticles; RBD, receptor binding domain; RBM, receptor binding motif.