Fig. 3.
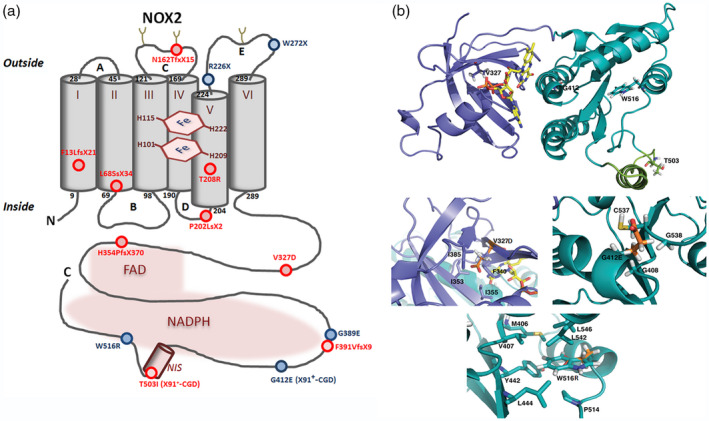
(a) Schematic representation of X‐linked chronic granulomatous disease (CGD) point mutations within the NOX2 protein – new mutations are shown in red and known mutations in blue. New mutations in the promoter (patient P1) or in intron sequences (patients P4a and P12) of CYBB are not represented. (b) NOX2 dehydrogenase model and modeling of CGD mutations. The DH model is represented as ribbons, dark green for the nicotinamide adenine dinucleotide phosphate (NADPH) oxidase 2 (NOX2) binding domain – corresponding to the 3A1F Protein Data Bank (db) structure – and purple‐blue for the modelled FAD binding domain ([37]). NOX2 NIS, in light green, is an approximate structural model but is partly disordered, flexible as suggested by the absence of density in the 3A1F structure for this sequence. Some of the residues corresponding to X910 and X91+ mutations are highlighted as shown in stick (V327, G412, W516 and T503). Three zooms highlight the V327, G412 and W516 residues with emphasis on the neighboring residues around the considered mutations. In each case, the mutated position is presented with both the wild‐type side chain and the corresponding mutation, in orange, superimposed. Molecular graphic images were produced using PyMOL software
