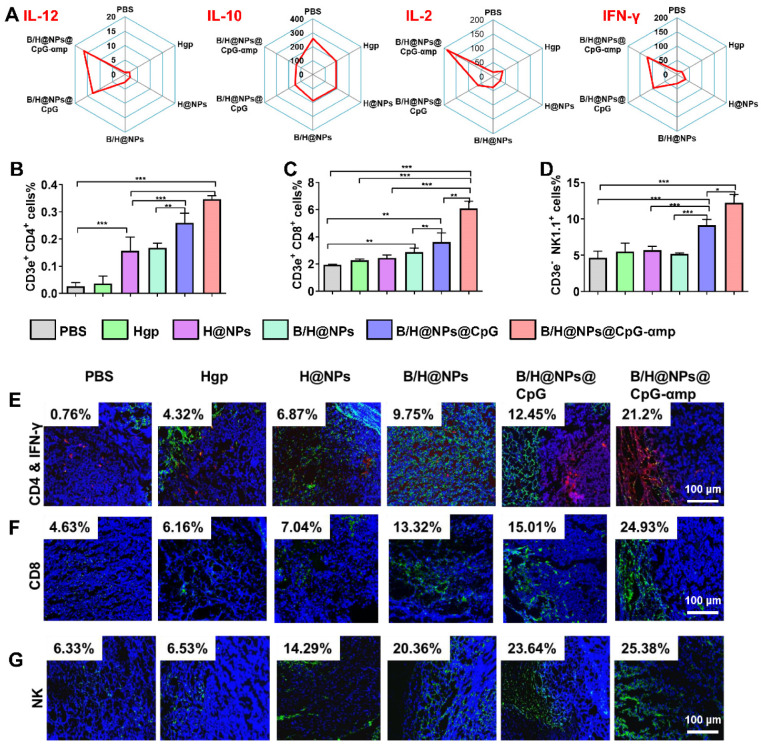
Figure 6.
Different nano-complexes remodel the tumor microenvironment in B16 tumor-bearing mice. (A) The expression of IL-12, IL-2, IFN-γ and IL-10 cytokines at the tumor site were analyzed using an ELISA kit. (B, C, D) The activation of Th1 cells (CD4+ T), cytotoxic T cells (CD8+ T), and natural killer cells (NK) in tumors were examined via flow cytometry. (E, F, G) The infiltration of Th1 (CD4+/IFN-γ), CD8+, and NK cells at tumor sites examined by immunofluorescent staining. Blue: nucleus, Red: CD4+ cells, Green: IFN-γ, CD8+ cells, and NK cells, Yellow: the co-localization of CD4+ cells and IFN-γ. The data were analyzed by automatic multispectral imaging system (PerkinElmer Vectra II). Scale bar:100 μm. For E-G, the quantifications are shown in Figure S9A-C. Three mice were analyzed in every group (n = 3), and one representative image per group are displayed. Data are the mean ± SEM and representative of three independent experiments. Differences between two groups were tested using an unpaired, two-tailed Student's t-test. Differences among multiple groups were tested with one-way ANOVA followed by Tukey's multiple comparison. Significant differences between groups are expressed as follows: *P < 0.05, **P < 0.01, or ***P < 0.001.