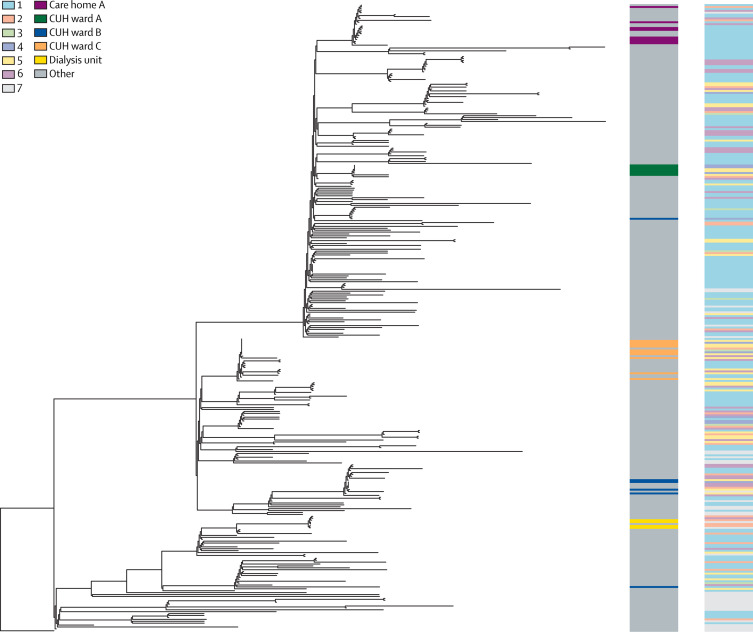
Figure 4.
Phylogenetic tree of SARS-CoV-2 genomes from CUH
Phylogenetic tree of 299 SARS-CoV-2 genomes form CUH and 30 international genomes. The tree is rooted on a December, 2019, genome from Wuhan, China. The left-hand column highlights several hospital and community associated clusters in different colours. Wards A, B, and C all had clusters of hospital-acquired infections cases with viruses of less than two single nucleotide polymorphisms different. Eight ward C cases are contained within one of the largest clusters of identical viruses. Genomic clusters containing the cases for wards B and C, the dialysis unit, and care home A all included health-care workers. The right hand column shows the classification of infection, also shown in figure 2. Classification of infection: (1) community onset, community associated; (2) community onset, suspected health-care associated; (3) hospital onset, indeterminate health-care associated; (4) hospital onset, suspected health-care associated; (5) hospital onset, health-care associated; (6) health-care worker; (7) unable to determine or missing. See appendix pp 23–24 for GISAID identification codes of included reference genomes, and appendix p 13 for their position on the East of England phylogenetic tree. SARS-CoV-2=severe acute respiratory syndrome coronavirus 2. CUH=Cambridge University Hospitals National Health Service Foundation Trust.