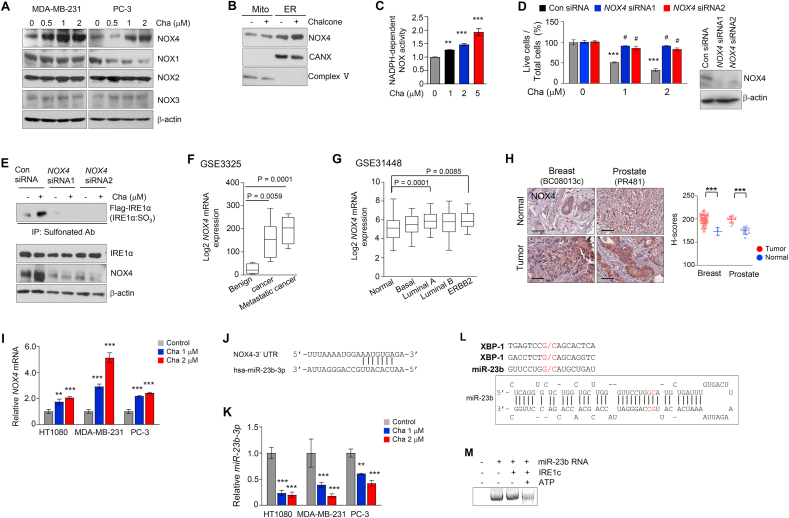
Fig. 3.
Degradation of miR-23b by IRE1α-RIDD increased NOX4 expression in ER. (A) Immunoblotting of NOX family was performed using cell lysates treated with different concentrations of chalcone in MDA-MB-231 and PC-3. (B) Immunoblotting of NOX4 was performed in subcellular fractionation of PC-3 cells treated with chalcone. (C) NOX activity was measured by chemiluminescence (n = 3 independent experiments). (D) Live cells was confirmed in PC-3 cells with NOX4 siRNA transfection compared with cells transfected with scrambled siRNA oligonucleotides (n = 3 independent experiments). Right; immunoblotting was performed to validate knockdown of NOX4. (E) Immunoprecipitation using anti-sulfonate antibody was performed on MDA-MB-231 cells with NOX4 siRNA, scrambled siRNA upon the treatment of chalcone. (F-G) NOX4 expression was analyzed using the Gene Expression Omnibus database from NCBI. Prostate (GSE3325, F), and Breast (GSE31448, G) datasets are presented. (H) Representative immunohistochemical staining of NOX4 on tissue microarrays. Right; quantification data of NOX4 expression. Scale bar, 100 μm. (brown: positive antibody staining, blue: hematoxylin for nuclear staining). (I) The mRNA expression of NOX4 was measured by qRT-PCR (n = 3 independent experiments). (J) Sequence motif of miR-23b-3p binding sites to NOX4. (K) miR-23b-3p expression was detected after treatment of chalcone (n = 3 independent experiments). (L) Sequence motif for the IRE1α cleavage sites (upper) and predicted secondary structures of miR-23b with their potential IRE1α cleavage sites (G/C sites marked with red arrows). (M) In vitro IRE1α mediated miR-23b cleavage assay. In vitro–transcribed miR-23b RNA was incubated with or without recombinant IRE1α protein in the presence or absence of ATP in the reaction buffer. The cleavage reaction products were resolved on an agarose gel. Data represent mean ± SD. Statistical differences were detected with one-way ANOVA followed by Tukey post hoc test (C, F, G, H), and two-way ANOVA followed by Bonferroni's test (D, I, K). Asterisks indicate significant differences from the control. Hashtag indicates significant differences from the chalcone. Cha; chalcone. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)