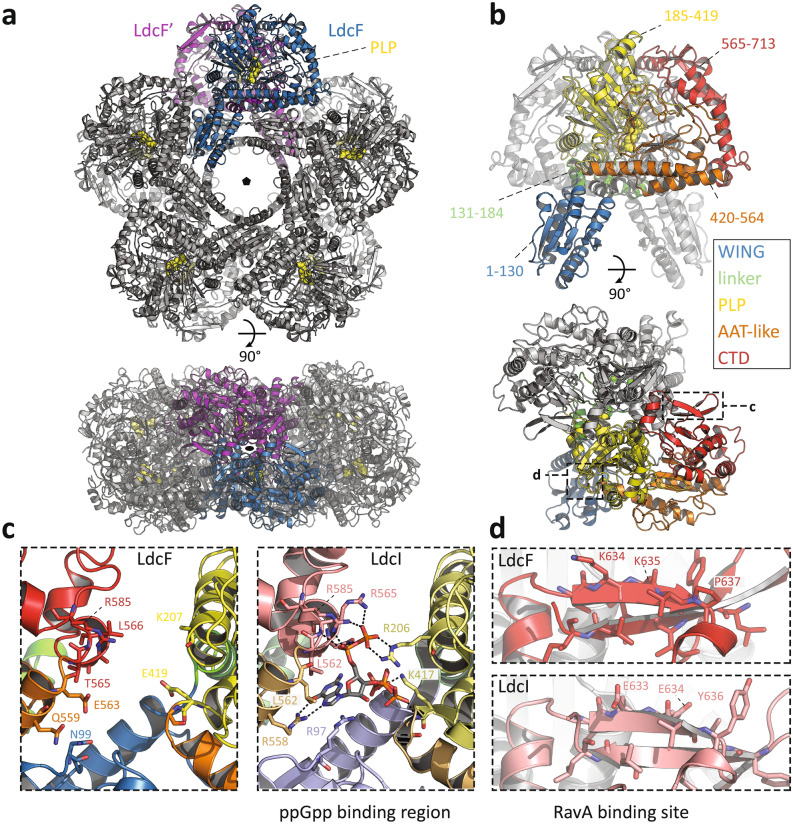
Figure 3.
Crystal structure of the F. novicida lysine decarboxylase LdcF. (a) Front (upper panel) and side view (lower panel) of decameric LdcF, with one highlighted dimer coloured blue and purple, while other dimers are coloured light and dark grey. The covalently bound pyridoxal phosphate (PLP) cofactor is shown as yellow spheres. (b) Front (upper panel) and side view (lower panel) of an LdcF dimer extracted from the decamer shown in (a). In one monomer, different domains are coloured according to a rainbow scheme (WING domain: blue, linker: green, PLP-binding domain: yellow, AAT-like domain: orange, C-terminal domain: red), with accompanying annotated amino acid residue ranges. (c) Comparison between the AAT-like domains (termed ppGpp binding domain in E. coli LdcI) of F. novicida LdcF (left) and E. coli LdcI (right). Residues of E. coli LdcI involved in ppGpp binding, and the corresponding residues in the AAT-like domain of F. novicida LdcF are annotated and shown as sticks. Domains are coloured as in (b), but using lighter tints for E. coli LdcI. (d) Comparison between the RavA-binding site in E. coli LdcI, and the corresponding region in F. novicida LdcF. Residues of E. coli LdcI involved in RavA binding, and the corresponding residues of F. novicida LdcF are annotated and shown as sticks.