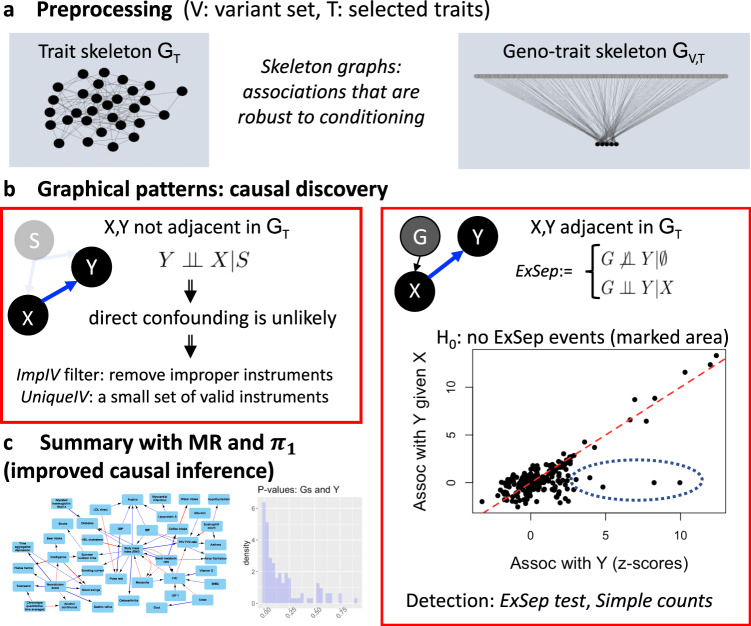
Fig. 1. The cGAUGE workflow for causal discovery.
We analyze genetic and phenotypic data collected from independent subjects. a We first preprocess the data to infer skeleton graphs: graphs that represent associations that are robust to conditioning. Based on causal inference theory1,2, surviving associations contain the subset of true causal links. We learn two skeletons: GT among the phenotypes (ignoring the genetic data in the process), and GV,T between the variants and the phenotypes. b We then analyze the edges and the non-edges of GT separately. We present methods that use GV,T to filter out improper instruments (ImpIV) or identify unique proper instruments (UniqueIV). While their theoretical justification is pertinent to GT non-edges, we illustrate using simulations how they reduce the empirical FDR when applied to all phenotype pairs. For GT edges we present an analysis based on ExSep events: associations between genetic variables and a trait Y that “disappear” once conditioned on a new phenotype X (i.e., p > p2). Under our local faithfulness assumption these patterns are evidence for a causal link from X to Y. The ExSep model selection test is a method to analyze all genetic variables under the null hypothesis of no ExSep events. c Finally, we utilize our results for improved inference using Mendelian Randomization (MR) and also utilize the π1 estimate for each exposure–outcome pair. This score quantifies the consistency of the associations between the exposure’s instruments and the outcome, which can be used to flag potential false positive causal links.