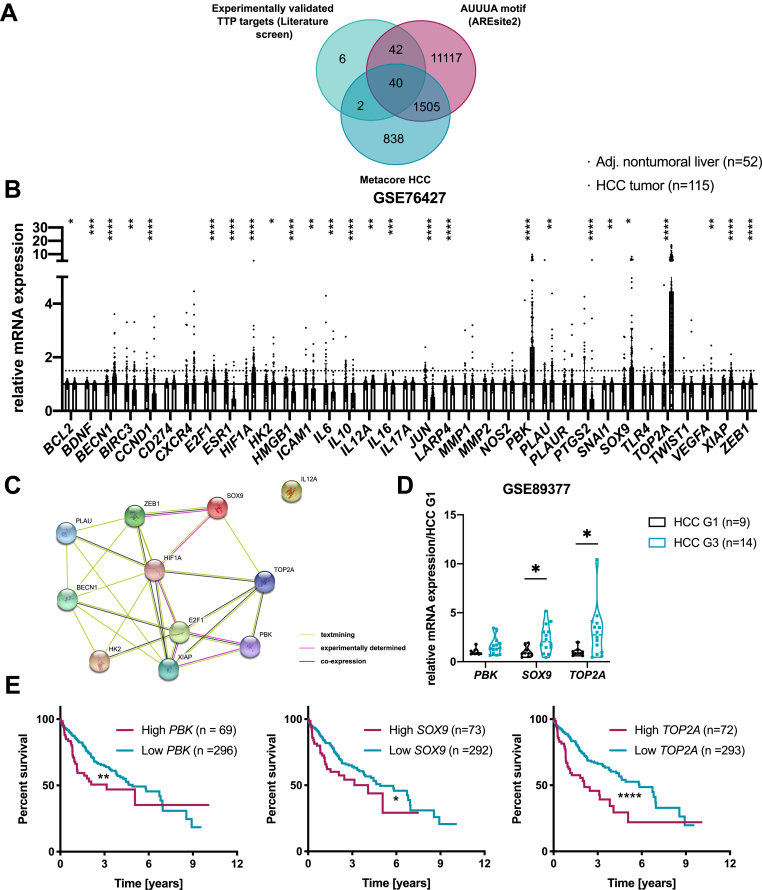
Figure 7.
Bioinformatic predictions of cancer-promoting factors potentially targeted by TTP in HCC. (A) Venn diagram showing TTP targets identified by crossing experimentally validated TTP targets with genes containing an AUUUA motif (based on AREsite2) and HCC-related factors (MetaCore analyses). (B) mRNA fold change of 40 potential targets in HCC tumors (n = 115) versus non-tumoral liver (n = 52) (GSE76427 transcriptomic dataset). (C) String analysis of interactions between 11 potential TTP targets (STRING database). (D) mRNA fold change of candidates up-regulated >1.5-fold in GSE76427 dataset from (B) (PBK, SOX9, and TOP2A) in HCC G1 (n = 9) and HCC G3 tumors (n = 14) of second human transcriptomic dataset (GSE89377). Data represent mean ± SD. (E) Survival analysis of HCC patients, patients stratified based on PBK, SOX9, and TOP2A mRNA expression levels (80th percentile). P value was calculated using a log-rank test (data retrieved from TCGA and Human Protein Atlas). For transcriptomics analyses of GEO datasets the P value was corrected for multiple testing using the Benjamini, Krieger and Yekutieli procedure (α = 5%). ∗∗∗P < .001, ∗∗P < .01, ∗P < .05.