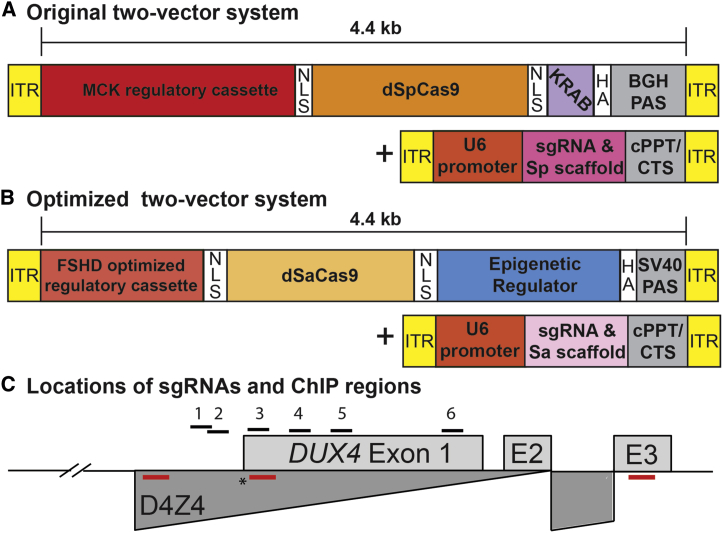
Figure 1.
CRISPRi constructs for epigenetic repression of DUX4
(A) Original two-vector system: (1) dSpCas9 fused to the KRAB TRD under control of the CK8 regulatory cassette,39,40 and (2) a DUX4-targeting sgRNA with dSpCas9-compatible scaffold under control of the U6 promoter. (B) Optimized two-vector system: (1) the smaller dSaCas9 ortholog fused to one of four epigenetic repressors (HP1α, HP1γ, the MeCP2 TRD, or the SUV39H1 pre-SET, SET, and post-SET domains) under control of a minimized skeletal muscle regulatory cassette, and (2) a DUX4-targeting sgRNA with dSaCas9-compatible scaffold incorporating modifications that remove a putative Pol III terminator and improve assembly with dCas941 under control of the U6 promoter. (C) Schematic diagram of the FSHD locus at chromosome 4q35. Distances are shown relative to the DUX4 MAL start codon (∗). For simplicity, only the distal D4Z4 repeat unit of the macrosatellite array (dark gray) is depicted. DUX4 exons 1 and 2 are located within the D4Z4 repeat, and exon 3 lies in the distal subtelomeric sequence. The locations of sgRNA target sequences (#1–6) are indicated. Positions of ChIP amplicons are shown as unlabeled red bars (in order from 5′ to 3′: DUX4 promoter, exon 1, and exon 3). Refer to text for additional details. ITR, inverted terminal repeat; NLS, nuclear localization signal; HGA, hemagglutinin; bGH, bovine growth hormone; PAS, polyadenylation signal; cPPT, central polypurine tract; CTS, central termination sequence.