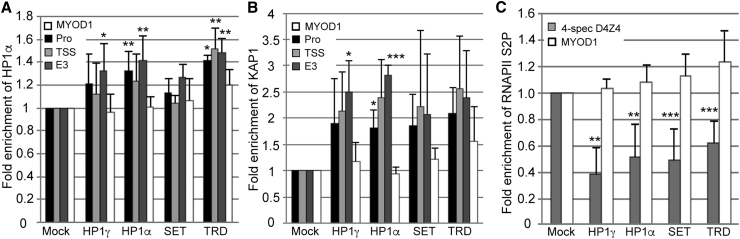
Figure 5.
Targeting dSaCas9-repressors to DUX4 increases chromatin repression at the locus
ChIP assays were performed using FSHD myocytes transduced with each dSaCas9-epigenetic regulator + individual optimal sgRNA targeting the DUX4 promoter or exon 1. (A–C) Chromatin was immunoprecipitated using antibodies specific for (A) HP1α or (B) KAP1 and analyzed by qPCR using primers to the promoter (Pro), transcription start site (TSS), or exon 3 of DUX4 or to MYOD1, or (C) antibodies specific for the elongating form of RNA Pol II (phospho-serine 2) and analyzed by qPCR using primers specific to DUX4 exon1/intron1 on chromosome 4 or to MYOD1. MYOD1 was used as a negative control for an active gene that should not be affected by CRISPRi targeted to DUX4. Locations of DUX4 primers are shown in Figure 1C. Data are presented as fold enrichment of the target region by each specific antibody normalized to α-histone H3, with enrichment for mock-infected cells set to 1. For all panels, each bar represents the average of at least three independent ChIP experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 are from comparing to enrichment at MYOD1.