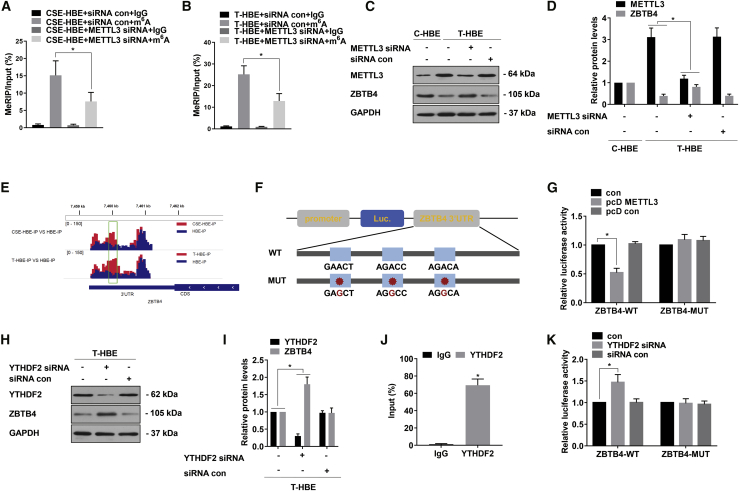
Figure 4.
Silencing of ZBTB4 by a METTL3-m6A-YTHDF2-dependent mechanism
T-HBE cells were transfected with METTL3 siRNA or siRNA con, pcD METTL3, or pcD con and YTHDF2 siRNA or siRNA con. (A) Methylated RNA immunoprecipitation (meRIP)-qPCR was applied to assess the m6A levels for ZBTB4 in CSE-HBE cells (mean ± SD, n = 3). (B) meRIP-qPCR was applied to assess the m6A levels for ZBTB4 in T-HBE cells (mean ± SD, n = 3). (C) Western blots were performed, and (D) relative protein levels (mean ± SD, n = 3) of METTL3 and ZBTB4 were determined. (E) Peaks indicating the relative abundance of m6A sites in ZBTB4 mRNA. (F) WT or m6A consensus sequence mutant ZBTB4 3′ UTR was fused with a firefly luciferase reporter. Mutations of m6A consensus sequences were generated by replacing A with G. (G) Relative activities of the WT and Mut luciferase reporters in T-HBE cells (mean ± SD, n = 3). (H) western blots were performed, and (I) relative protein levels (mean ± SD, n = 3) of YTHDF2 and ZBTB4 were determined. (J) RIP assays were performed in T-HBE cells to detect the direct binding between the ZBTB4 mRNA and YTHDF2 protein (mean ± SD, n = 3). (K) Relative activities of the WT and Mut luciferase reporters in T-HBE cells (mean ± SD, n = 3). ∗p < 0.05, different from control group.