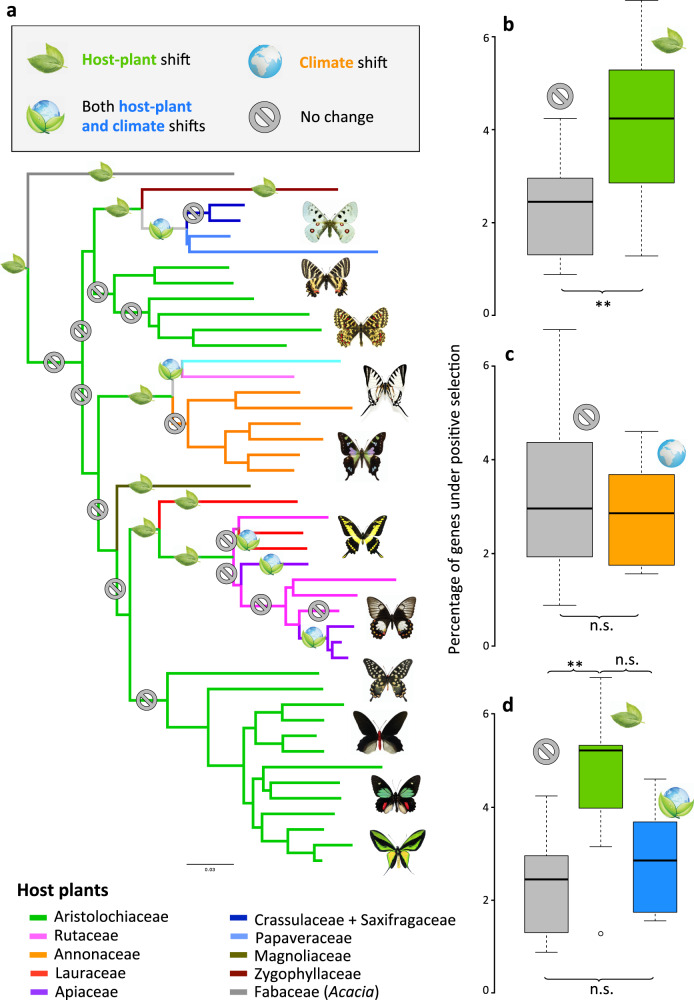
Fig. 4. Host–plant shifts promote higher molecular adaptations.
a Genus-level phylogenomic tree displaying branches with and without host–plant shifts, on which genome-wide analyses of molecular evolution are performed. b Number of genes under positive selection (dN/dS > 1) for swallowtail lineages shifting to new host–plant families (n = 14, green) or not (n = 14, grey). c Number of genes under positive selection for swallowtail lineages undergoing climate shifts (n = 5, orange) or not (n = 23, grey). d Number of genes under positive selection for swallowtail lineages shifting to new host plants (n = 9, green), shifting both host–plant and climate (n = 5, blue) or not (n = 14, grey). The proportion of genes was estimated with Dataset 2 (1533 genes, see Supplementary Fig. 19 for the results with Dataset 1 and 520 genes). This demonstrates genome-wide signatures of adaptations in swallowtail lineages shifting to new host–plant families. Genes under positive selection did not contain over- or under-represented functional GO categories (Supplementary Data 2). Wilcoxon rank-sum test: n.s. = not significant (P > 0.05), *P ≤ 0.05, **P ≤ 0.01. Pictures and icons made by Fabien Condamine.