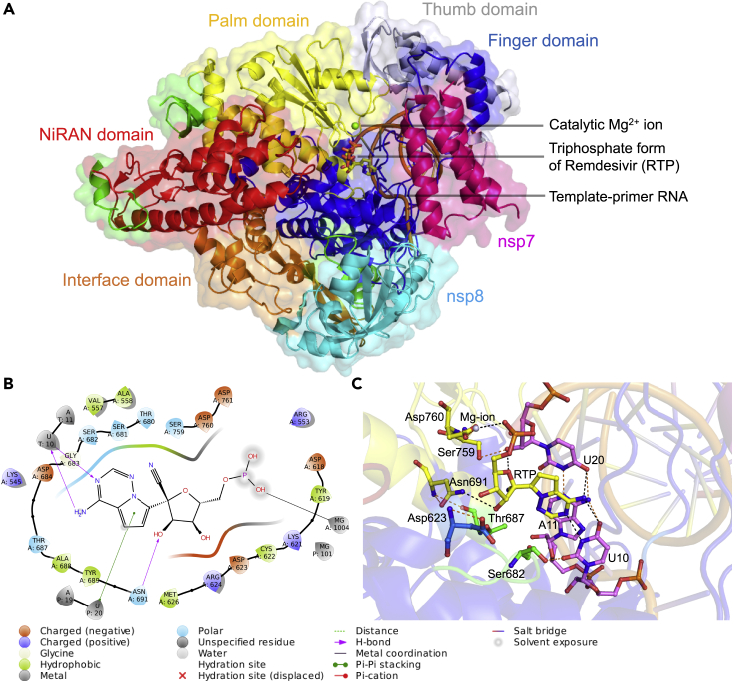
Figure 1.
Structure of the nsp12-nsp7-nsp8 complex bound to the template-primer RNA and triphosphate form of remdesivir (RTP) and the RTP-interacting residues
(A) The cryo-electron microscopy structure of the RNA-dependent RNA polymerase from SARS-CoV-2 (PDB ID: 7BV2) consisting of nsp12-nsp7-nsp8 complex bound to template-primer RNA, the triphosphate form of remdesivir (RTP), and the catalytic Mg2+ ion is shown. Here nsp7 and nps8 are shown in magenta and cyan colors, respectively. The nsp12 subunit is comprised of several subdomains, where the NiRAN domain from residues 115-250 is shown in red color and the interface domain from residues 251–365 is shown in orange color. The C-terminal comprising residues 366–920 is divided into three subdomains, where finger domain (residues 398–581 and 621–679), palm domain (residues 582–627 and 688–815), and thumb domain (residues 816–919) are shown in blue, yellow, and blue-white colors, respectively. The triphosphate form of remdesivir (RTP) is shown as a stick model.
(B) Ligand interaction diagram of remdesivir within a 6-Å distance of the nsp12-nsp7-nsp8 complex bound to the template-primer RNA is shown, and the types of intermolecular interactions are labeled.
(C) Interacting residues of corresponding nsp12 subdomains (green, blue, and yellow) and RNA bases (violet) of remdesivir (yellow) are labeled and shown as stick models. The Mg2+ (gray sphere) coordinating the RTP is shown. Hydrogen bond and salt bridge interactions are shown as black and orange dashes, respectively.