Figure 4.
Myc is a central component of the regulatory networks driving osteoclast-lineage commitment
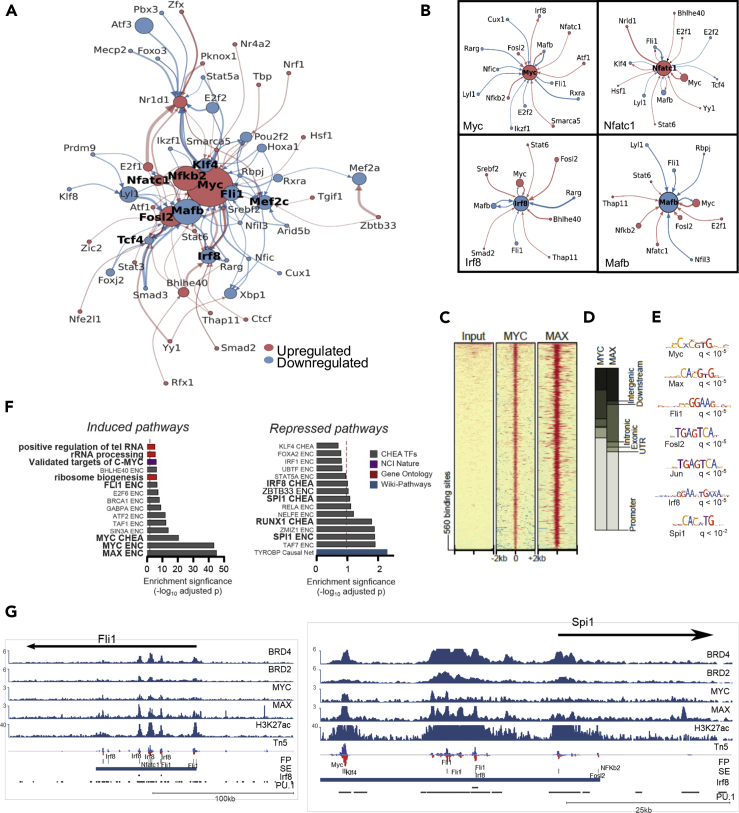
(A) Chromatin accessibility-based construction of gene regulatory network that drives transcriptional changes during OC lineage commitment. Directed TF network diagram: arrow width indicates footprint score for originating TFs at target gene promoter/enhancer, and color indicates direction of expression change for originating TFs (red: upregulated and blue: downregulated). Node size indicates strength of expression change (-log10(padj)∗|log2FC|) and color indicates direction of change at 14 hr after RANKL treatment.
(B) Individual TF modules comprising core components of the overall network.
(C–E) (C) Heatmap representation of ChIP-seq against Myc and Max, along with (D) genomic annotation of significantly enriched areas and (E) thier motif analysis.
(F) Overrepresentation analysis of direct Myc gene targets (as predicted upon integrated cistrome-transcriptome analysis) using EnrichR.
(G) IGV snapshots of Fli1 and Spi1 genetic loci. From top to bottom: showing Brd2/4, Myc, Max and H3K27Ac ChIP-seq, ATAC-seq and Pu.1. and Irf8 ChiP-seq are from (Langlais et al., 2016).