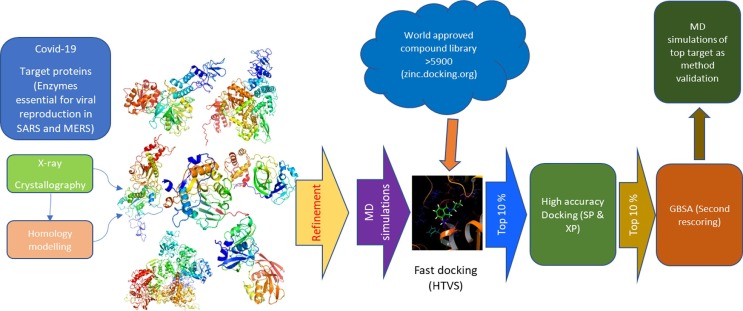
Fig 1.
Schematic road map of the overall study design. The protein models from various protein bank and other sources were optimized and relaxed by MD simulations. The relaxed structures were then mapped for active site and used to generate GLIDE Grid for HT-virtual screen with world approved drug libraries. The top 10% of these compounds were subjected to high accuracy docking (SP/XP) which were then further refined to the top 10%. This was followed by a secondary rescoring (GBSA). Top leads were subjected to MD simulations of the top compounds for each viral target tested as a methodological validation.