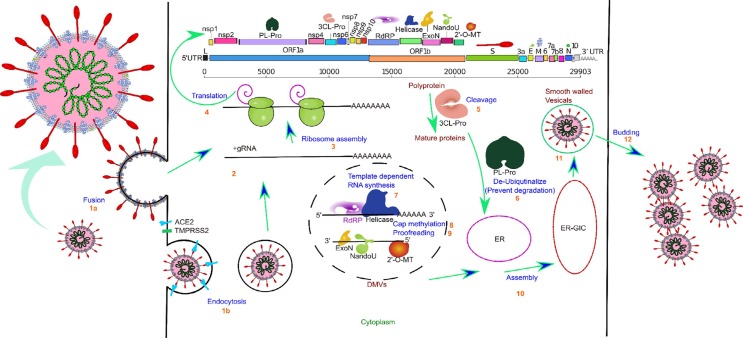
Fig. 2.
Graphical illustration of the SARS-CoV-2 life cycle along with target proteins used in HTVS. The infection cycle starts when the SARS-CoV-2 Spike protein binds to the Human ACE2 receptor. An S1-induced post-stable S2 conformation allows either viral-host cell fusion (1a) or endocytosis (1b). Fusion directly allows the viral RNA to enter the host cell (2), but endocytosis requires lysosomal degradation of coat and envelop for the release of viral nucleocapsid in the cytoplasm. The large viral script is known to encode 29 viral proteins (3), including the 7 essential nonstructural proteins that are selected as targets in our paper. A replicase is used to translate most of the viral genomic RNA to synthesize two replicase polyproteins, pp1a and pp1ab, and many small ORFs(4). The two major polyproteins are processed by two proteases, PLpro and 3CLpro(5), generating 16 nonstructural proteins. ExoN possesses a viral exoribonuclease activity that acts on both ssRNA and dsRNA in a 3′ to 5′ direction(9). Viral Helicase plays a critical role in viral replication by expediting appropriate folding (7). The enzyme 2′-O-MT methylates the viral 2′ end which is important for the virion to avoid host recognition of their RNA (8). RdRP is involved in viral-host cell replication through catalyzing template synthesis of polynucleotides in the 5′ to 3′ direction (7). NendoU is an Mn2+ dependent hexamer (dimer of trimer) enzyme with sparse functional information. The most prominent theory regarding NendoU is that the activity of this protein is responsible for protein interference with the innate immune system. For viral assembly of S, E, and M proteins in the endoplasmic reticulum, along with the N protein are combined with the (+) gRNA to become a helical nucleoprotein complex. They assemble to form a virus particle in the endoplasmic reticulum-Golgi apparatus compartment, this particle is then excreted from the cell through budding mediated by fusion of smooth-walled vesicles to the plasma membrane (11–12).