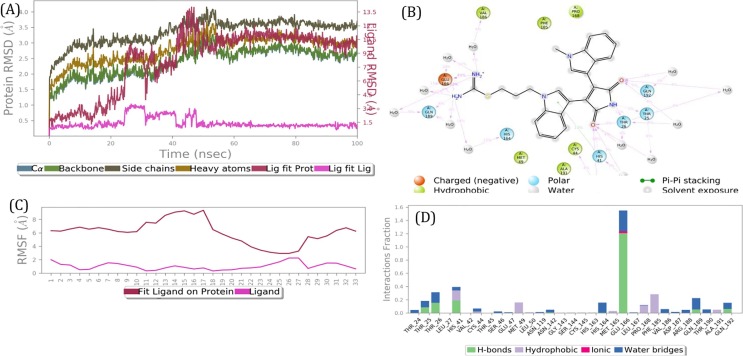
Fig. 3.
Results of a 100 nano seconds (ns) MD simulation. (A) Root mean square deviations difference between the Main protease (3CLPro) and bound ligand BIM IX (<4 Å). Graph obtained for RMSF value of ligand (purple line) from the protein back bone (green line). It revealed that there was a major conformational change of the ligand at around 50 ns without loss of the ligand. This suggests two binding conformers of same ligand within the binding site. (B) Schematic 2D representation of bound ligand interactions of BIM IX throughout the simulation. (C) Root mean square fluctuation between the binding site of target protein and interacting ligand. (D) Critical protein ligand contacts of amino acid side chain residues with the interaction properties. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)