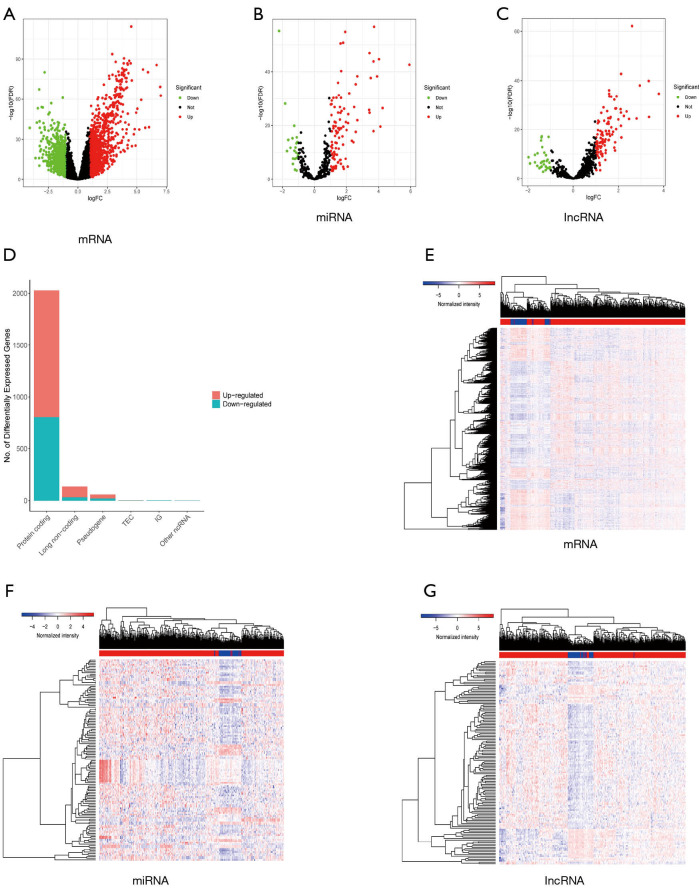
Figure 2.
Identification of differentially expressed RNAs. (A,B,C) The volcano plots of differentially expressed mRNAs (A), miRNAs (B), and lncRNAs between normal and tumor samples. (A,B,C) We defined the absolute value of logFC >1 and the FDR value <0.05 as the cutoff value. Red points represent upregulated RNAs, green dots represent downregulated RNAs, and black points represent the unchanged RNAs which did not meet the criteria. (D) The bar plot illustrates the differentially expressed RNAs among which 222 mRNAs, 107 miRNAs, and 104 lncRNAs were upregulated (red bar), while 806 mRNAs, 21 miRNAs, and 32 lncRNAs were downregulated (blue bar). (E,F,G) The heatmap also shows the differentially expressed RNAs between adjacent and tumorous tissues. mRNA, messenger RNA; miRNA, microRNA; lncRNA, long noncoding RNA; FDR, false discovery rate.