Abstract
Background
Equine gut microbiology studies to date have primarily focused on horses and ponies, which represent only one of the eight extant equine species. This is despite asses and mules comprising almost half of the world’s domesticated equines, and donkeys being superior to horses/ponies in their ability to degrade dietary fiber. Limited attention has also been given to commensal anaerobic fungi and archaea even though anaerobic fungi are potent fiber degrading organisms, the activity of which is enhanced by methanogenic archaea. Therefore, the objective of this study was to broaden the current knowledge of bacterial, anaerobic fungal and archaeal diversity of the equine fecal microbiota to multiple species of equines. Core taxa shared by all the equine fecal samples (n = 70) were determined and an overview given of the microbiota across different equine types (horse, donkey, horse × donkey and zebra).
Results
Equine type was associated with differences in both fecal microbial concentrations and community composition. Donkey was generally most distinct from the other equine types, with horse and zebra not differing. Despite this, a common bacterial core of eight OTUs (out of 2070) and 16 genus level groupings (out of 231) was found in all the fecal samples. This bacterial core represented a much larger proportion of the equine fecal microbiota than previously reported, primarily due to the detection of predominant core taxa belonging to the phyla Kiritimatiellaeota (formerly Verrucomicrobia subdivision 5) and Spirochaetes. The majority of the core bacterial taxa lack cultured representation. Archaea and anaerobic fungi were present in all animals, however, no core taxon was detected for either despite several taxa being prevalent and predominant.
Conclusions
Whilst differences were observed between equine types, a core fecal microbiota existed across all the equines. This core was composed primarily of a few predominant bacterial taxa, the majority of which are novel and lack cultured representation. The lack of microbial cultures representing the predominant taxa needs to be addressed, as their availability is essential to gain fundamental knowledge of the microbial functions that underpin the equine hindgut ecosystem.
Keywords: Feces, Barcoded amplicon sequencing, Bacteria, Archaea, Anaerobic fungi, Horse, Donkey, Mule, Hinny, Zebra
Background
The hindgut microbiome is key to the ability of equines to degrade dietary fiber, as equines themselves lack fiber degrading enzymes. In recent years, there has been a move to characterizing the equine hindgut microbiota using high throughput sequencing of fecal [1–7] and digesta samples [8, 9], as well as determining the equine fecal and hindgut core composition [3, 7, 8, 10, 11]. This is particularly important considering the variation reported in the hindgut microbiota and metabolome of different animals [12]. By determining the core microbiota of healthy equines, insights can be gained into cornerstone taxa and functions present in the ecosystem [13, 14].
Whilst the equine hindgut bacterial core has been described, its composition differs between studies [7, 8, 10, 11]. To date, it has been concluded that diet changes the composition and size of the equine bacterial core [10], unlike age and obesity which have minimal effects [7, 10]. The bacterial core has also been shown to be represented by multiple low abundance taxa, which together are of small cumulative abundance [7, 8, 10]. However, studies of the bacterial core to date have focused only on domesticated horses and/or ponies, which represent only one of the eight extant species of the equid family [15].
The knowledge of the gut microbiota from horses and ponies is normally directly translated to donkeys and mules, which comprise 47.9% of the world’s 113 million domesticated equines [16]. The validity of this direct translation is not clear particularly as diet and gut transit time, which are key influencing factors of the hindgut microbiome, differ between horses/ponies and donkeys [17, 18]. Differences also occur in feed digestibility, with donkeys relative to ponies consistently having a higher dry matter digestibility for a given diet [18].
Furthermore, bacteria are not alone in the equine hindgut, as anaerobic fungi, methanogenic archaea and protozoa are also present [19]. Bacteria as well as anaerobic fungi are the primary degraders of fiber in mammalian herbivores. However, anaerobic fungi are significantly better at degrading plant cell walls than bacteria [20], due to their invasive growth and potent fiber degrading enzymes [21, 22]. Methanogenic archaea do not degrade fiber, but are known to promote anaerobic fungal activity due to the removal of fermentation end products [23]. Protozoa have been suggested not to play a major role in hindgut fiber degradation, as cellulose digestion is not changed by their removal [24].
The majority of knowledge on anaerobic fungi and methanogenic archaea in the gut of mammalian herbivores is based on ruminants, with only limited information on equines [19]. There is evidence that equine anaerobic fungi are distinct from those found in ruminants, both in terms of taxonomy and physiology [25, 26]. In terms of methanogenic archaea, sequencing based studies of fecal samples have indicated that the genera Methanocorpusculum and Methanobrevibacter predominate [6, 27]. It remains to be verified, however, if these two genera are both part of the core hindgut microbiota of equines.
Protozoa present in the equine hindgut are similar to those present in the rumen of ruminants in terms of being dominated by ciliates, although amoeboid and flagellated protozoa may also be present [19]. The majority of the 24 ciliate genera that have been described in equines to date, however, are not typically found in ruminants. The two ciliate protozoal genera Blepharocorys and Cycloposthium are thought to be the most prevalent in equines [19]. In recent years, two studies have used molecular methods based on rumen protozoal derived 18S rRNA gene primers to look at protozoal concentrations and diversity in the equine hindgut [6, 28]. However, caution is needed as the suitability of these rumen protozoal derived primers for study of equine ciliate protozoa remains to be verified. This can only be done once 18S rRNA gene sequences for the numerous equine protozoal genera described to date are available. As such, analysis of equine protozoa was not performed as part of this multi-kingdom study.
The objective of this study was, therefore, to determine the bacterial, archaeal and anaerobic fungal composition of fecal samples from a large cohort of equines (n = 70) that included five extant species (i.e. Equus ferus, Equus africanus, Equus quagga, Equus zebra and Equus greyvii), as well as mules and hinnies (i.e. horse × donkey). All animals were fed pasture or hay/haylage based diets and, in some cases, received complementary feedstuffs (< 1 kg/day) in order to meet dietary requirements (Additional file 1: Table S1). In order to capture as much variation as possible, the animals were sourced from multiple geographical locations. From this dataset an overview of the microbiota across equine types is given, followed by identification of the core bacteria, anaerobic fungi and archaea.
Results
The equine cohort (n = 70) studied here was composed of healthy animals aged between 4 and 26 years that had no known history of any gut mediated disease. The cohort included horses and ponies (Equus ferus caballus, n = 18), donkeys (Equus africanus asinus, n = 18), hybrids of donkey and horse (Equus ferus caballus × Equus africanus asinus, n = 18) and several different (sub)species of zebras (Equus quagga burchellii, n = 2; Equus quagga boehmi, n = 8; Equus zebra hartmannae n = 3; Equus greyvii, n = 3). Details of the individual animals including their location, diet and management are given in Additional file 1: Table S1. For the purpose of providing an overview of the fecal microbiota across equine types, the animals were classed as being either horse (n = 18), donkey (n = 18), horse × donkey (n = 18) or zebra (n = 16).
Microbial concentrations
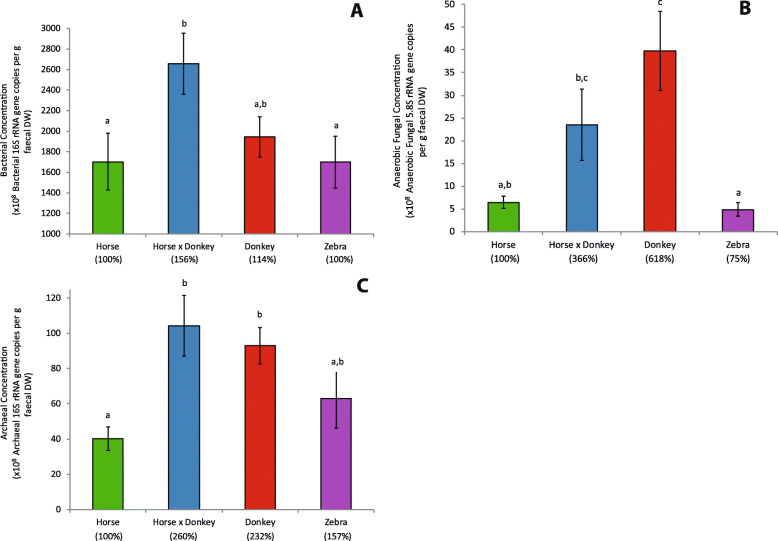
Due to equine type being associated with differences in fecal dry matter content (P = 0.006; Additional file 2: Figure S1), where zebra fecal dry matter was significantly higher than that of horse and of donkey, microbial concentration data were analyzed on a dry weight basis. Equine type was associated with differences in fecal bacterial concentrations (P = 0.016), with horse and zebra both being significantly lower than horse × donkey, and donkey being intermediate (Fig. 1a). Equine type was also associated with fecal anaerobic fungal concentrations (P < 0.001), with donkey having over 6-fold and 8-fold higher concentrations than horse and zebra, respectively (Fig. 1b). Horse × donkey had an almost 5-fold higher anaerobic fungal concentration than zebra. Fecal archaeal concentrations were also associated with equine type (P < 0.001), with donkey and horse × donkey having significantly higher archaeal concentrations than horse, and zebra not significantly differing from any of the other equine types (Fig. 1c). Analysis of the same data on a fresh weight basis showed generally similar trends (Additional file 3: Figure S2).
Fig. 1.
Effect of equine type on the fecal bacterial (a), anaerobic fungal (b) and archaeal (c) concentrations on a dry weight basis. Columns represent the mean (n = 18, except for zebra where n = 16) and error bars the SEM. Letters above the bars within each plot indicate significant differences (P < 0.05). Percentages stated on the x-axis in brackets indicate how the mean of each equine type compared to that of the horse.
Prokaryotic community composition
The bacteria (mean ± standard deviation (SD): 96.2 ± 3.08% of the 16S rRNA gene sequences) were represented by 2070 different OTUs (operational taxonomic units) which could be summarized to 231 different genus-level phylogenetic groupings. The archaea (3.8 ± 3.08% of the 16S rRNA sequences) were represented by 48 different OTUs, which could be summarized into seven different genus-level phylogenetic groupings. Of the 17 phyla detected, the following six were the most predominant: Firmicutes, Bacteroidetes, Verrucomicrobia, Spirochaetes, Fibrobacteres and Euryarchaeota (Additional file 4: Figure S3). Of the 76 families that could be classified, the most predominant families included the Lachnospiraceae, Ruminococcaceae, Bacteroidales S24 − 7_group, Spirochaetaceae and Fibrobacteraceae (Additional file 5: Figure S4).
Differences in fecal prokaryotic alpha diversity was associated with equine type in terms of both the number of observed OTUs (P = 0.007) and Phylogenetic Diversity (P = 0.029). Donkey fecal microbiota had a lower number of observed OTUs (228 ± 28 OTUs) compared to that of horse (253 ± 28 OTUs), horse × donkey (258 ± 28 OTUs) and zebra (255 ± 26 OTUs). Donkey fecal microbiota also had a lower Phylogenetic Diversity (18.96 ± 1.142) compared to that of zebras (20.19 ± 1.032), with neither donkey nor zebra significantly differing from horse (19.34 ± 1.396) or horse × donkey (19.43 ± 1.097).
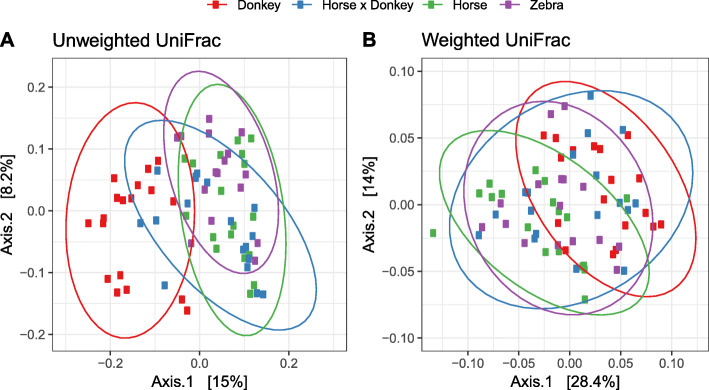
Beta diversity analysis of the fecal microbiota was performed using a non-constrained principal co-ordinate analysis (PCoA) at the OTU level based on pairwise UniFrac distances. Donkey fecal microbiota separated from the other equine types in the unweighted analysis but not the weighted analysis (Fig. 2). This suggests that the largest variation in the dataset was associated with donkey specific taxa occurring at a low relative abundance.
Fig. 2.
Unweighted (a) and weighted (b) UniFrac based principal co-ordinates analysis of the fecal prokaryotic community composition of the different equine types at the OTU level. Analysis used Log10 transformed data with ellipses representing 95% confidence intervals, and the percentages values labelled on each axis indicating the amount of total variation represented.
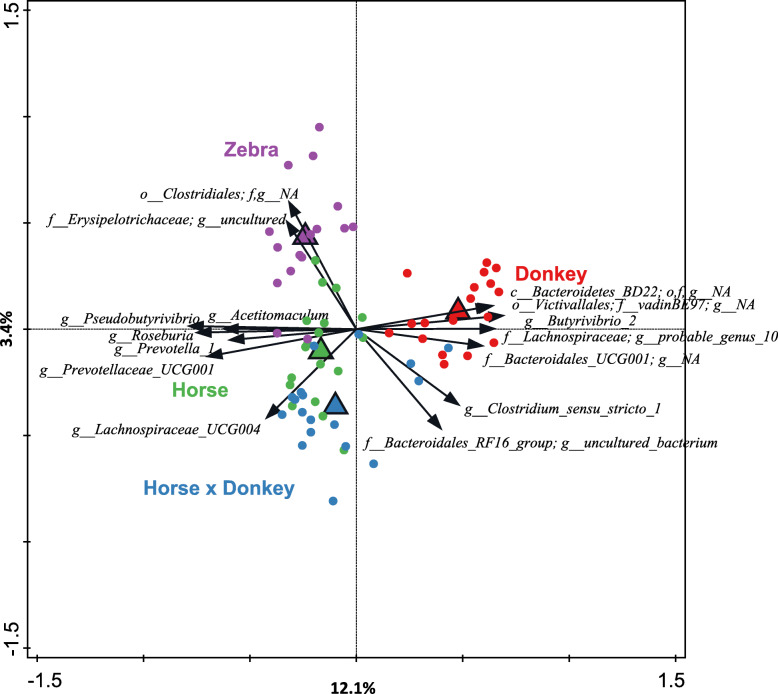
Redundancy analysis using genus-level phylogenetic groups confirmed that equine type was associated with differences in the prokaryotic community composition (P = 0.002), with equine type accounting for 18.3% of the total variation in the dataset (Fig. 3). The majority of the variation was represented by the first canonical axis, which showed that the prokaryotic community composition of donkey fecal microbiota separated from that of the three other equine types, the latter separating along the second canonical axis.
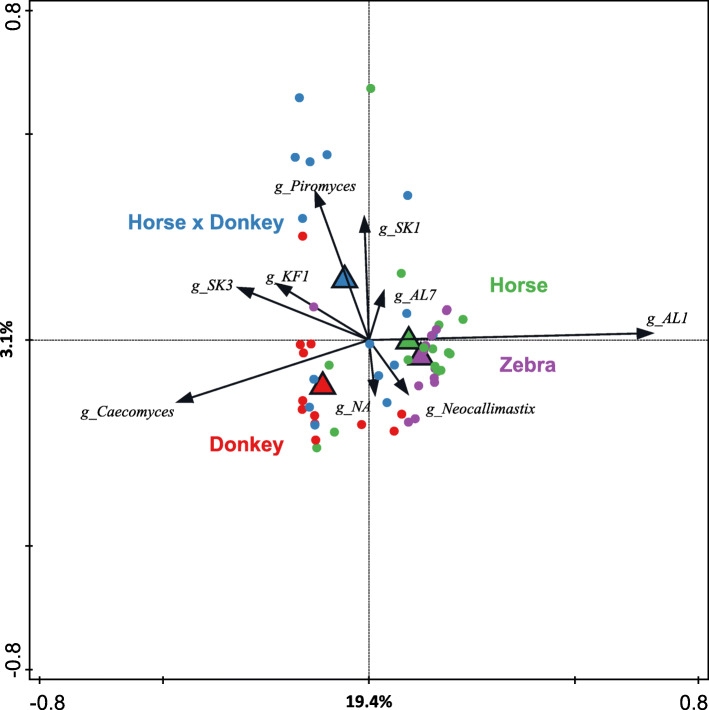
Fig. 3.
Redundancy analysis triplot showing the relationship between the top fifteen prokaryotic genus-level phylogenetic groupings of the OTUs for which the variation is best explained by the constrained axes. Arrow length indicates the variance that can be explained by equine type, with the perpendicular distance of the equine types to the arrow indicating the relative abundance of the genus-level phylogenetic grouping. Arrow labels indicate the taxonomic affiliation of genus-level phylogenetic groups, with the level (i.e. class (c), order (o), family (f) or genus (g)) and taxon (as defined by the Silva 16S rRNA database) that the groups could be reliably assigned to. For example ‘g_Prevotella_1’ represents an OTU reliably assigned to the Prevotella_1 genus, whereas “c_Bacteroidetes_BD2–2; o,f,g_NA” was reliably assigned to the class Bacteroidetes_BD2–2 but the order, family and genus could not be annotated (NA). Triangular symbols indicate the equine type means and circle symbols the individual samples color coded by equine type. Equine type explained 18.3% of the total variation in the dataset, and the plot axis are labelled with the amount of this they represent.
Five genus-level phylogenetic groups appeared to be positively associated with donkey. Of these five, two could be annotated to the genus level (Butyrivibrio 2, Lachnospiraceae probable_genus_10) and the others only to family (Bacteroidales UCG-001; Victivallales family_vadinBE97) or class level (Bacteroidetes BD2–2). Several genera appeared to generally be positively associated with horse, zebra and horse × donkey (and conversely negatively associated with donkey). These included Pseudobutyrivibrio, Roseburia, Prevotella 1, Acetitomaculum and Prevotellaceae UCG-001. The genus Lachnospiraceae UCG-004 appeared to be positively associated with horse × donkey and horse to different extents. An uncultured genus from the family Erysipelotrichaceae and an unknown family within the order Clostridiales appeared to be positively associated with zebra.
Anaerobic fungal community composition
For the analysis of anaerobic fungal community composition, reproducible PCR products of sufficient amount for sequencing were only obtained from 64 of the 70 different animals. The three horse and three zebra samples that failed (H4, H6, H7, Z11, Z14 & Z16: Additional file 1: Table S1) had the lowest anaerobic fungal concentrations detected in the cohort. Therefore, these six samples were considered to be below the detection limit of the amplicon sequencing method used in our study. From the 64 samples there were 358 OTUs detected, and these could be summarized to eight different genera. The taxonomic framework used in the anaerobic fungal ITS1 (AF-ITS1) database also includes as yet uncharacterized genus- or species-level clades [29]. Whilst the 358 OTUs could be summarized to nine clades, no further analysis at the clade level was performed as 53% of the OTUs could not be annotated at this level (Additional file 6: Figure S5). Of the eight genera detected that could be annotated (AL1, AL7, Caecomyces, KF1, Neocallimastix, Piromyces, SK1 and SK3), Caecomyces and the uncultivated genus AL1 predominated the dataset overall (Additional file 7: Figure S6). The other six genera were predominant and/or present in only a few individual animals (Additional file 7: Figure S6).
Differences in fecal anaerobic fungal alpha diversity was associated with equine type in terms of the number observed OTUs (P = 0.006), but not Phylogenetic Diversity (P = 0.989). Horse had a higher number of observed OTUs (22 ± 5 OTUs) compared to both donkey (16 ± 5 OTUs) and horse × donkey (16 ± 6 OTUs), with zebra (20 ± 6 OTUs) not differing from any of the other equine types.
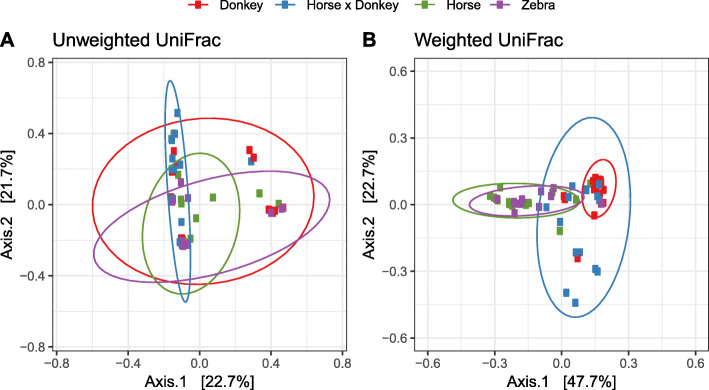
Beta diversity analysis of the fecal anaerobic fungal community using PCoA showed that all the equine types overlapped to some extent in the unweighted analysis (Fig. 4a). However, in the weighted PCoA the horse and zebra sample groups clustered separately from the donkey group and most variation was seen in the horse × donkey sample group (Fig. 4b).
Fig. 4.
Unweighted (a) and weighted (b) UniFrac based principal co-ordinates analysis of the fecal anaerobic fungal community composition of the different equine types at the OTU level. Analysis used Log10 transformed data with ellipses representing 95% confidence intervals, and the percentages values labelled on each axis indicating the amount of total variation represented.
Redundancy analysis using genus-level phylogenetic groups confirmed that equine type was associated with differences in the anaerobic fungal community composition (P = 0.002) and accounted for 23.6% of the total variation in the dataset (Fig. 5). Horse and zebra were positively associated with the genus AL1, whereas donkey was most positively associated with Caecomyces and the horse × donkey with Piromyces and SK1.
Fig. 5.
Redundancy analysis triplot showing the relationship between the anaerobic fungal genus-level phylogenetic groupings of the OTUs for which the variation is best explained by the constrained axes. Arrow length indicates the variance that can be explained by equine type, with the perpendicular distance of the equine types to the arrow indicating the relative abundance of the genus-level phylogenetic grouping. Arrow labels indicate the taxonomic affiliation that the genera could be reliably assigned to. For example ‘g_AL1’ represents a grouping reliably assigned to the AL1 genus, whereas ‘g_NA’ indicates that it was reliably assigned to the family Neocallimastigaceae but the genus could not be annotated (NA). Triangular symbols indicate the equine type means and circle symbols the individual samples color coded by equine type. Equine type explained 23.6% of the total variation in the dataset, and the plot axis are labelled with the amount of this they represent.
Equine Core microbiota analysis
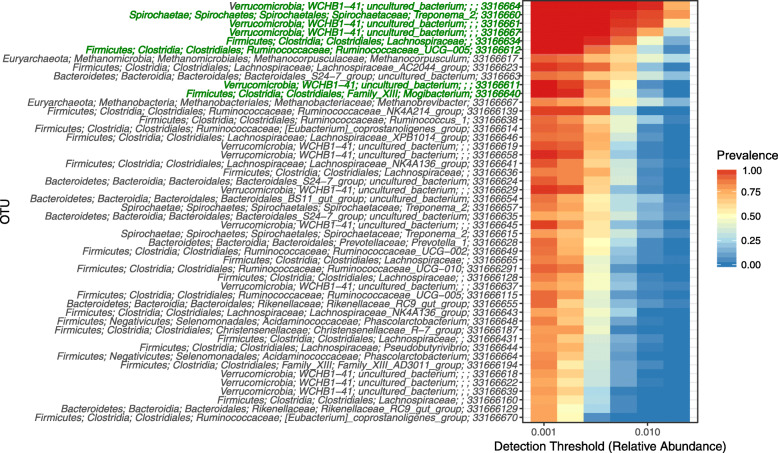
Analysis of the prokaryotic community composition at the OTU level indicated that of the 2118 OTUs detected, only 48 OTUs were present in at least 75% of the animals when a cut-off of > 0.001 was used (Fig. 6). Of these 48 OTUs, only eight OTUs were core, i.e. found in every animal (Fig. 6). Four of these OTUs belonged to an uncharacterized class, WCHB1–41, in the Verrucomicrobia phylum. The other four had annotation at lower taxonomic ranks: a non-annotated genus within the Lachnospiraceae, an uncharacterized genus called Ruminococcaceae UCG-005 and the two characterized genera Mogibacterium and Treponema 2. WCHB1–41 and Treponema 2 were the most abundant of the eight core OTUs (Table 1). These eight core OTUs represented 13.4% ± 3.26 (mean ± SD) of the prokaryotic community.
Fig. 6.
Heat map showing the relative abundance (> 0.001 cut-off) and prevalence (75% cut-off) of the prokaryotic OTUs in the 70 equine fecal samples analyzed. Different detection thresholds are used, providing information regarding the relative abundance of the OTUs relative to their prevalence. Taxonomic assignments of the OTUs are given to five taxonomic ranks (phylum, class, order, family and genus) where possible, followed by the OTU ID number. Where this was not possible, the non-annotated ranks were left empty (e.g. Verrucomicrobia; WCHB1–41; uncultured_bacterium;;; 3316664 has no information for the family and genus ranks). OTUs present in all animals (i.e. core) have their taxonomic assignments written in green.
Table 1.
Taxonomic annotationa and percentage relative abundancesb of the core taxa detected at the OTU level
| Phylum | Class | Order | Family | Genus | OTU No. | Mean | SD | SEM |
|---|---|---|---|---|---|---|---|---|
| Spirochaetae | Spirochaetes | Spirochaetales | Spirochaetaceae | Treponema 2 | 3316660 | 3.41 | 1.74 | 0.21 |
| Verrucomicrobia | WCHB1–41 | uncultured bacterium | NA | NA | 3316664 | 2.76 | 1.26 | 0.15 |
| Verrucomicrobia | WCHB1–41 | uncultured bacterium | NA | NA | 3316661 | 2.26 | 1.16 | 0.14 |
| Verrucomicrobia | WCHB1–41 | uncultured bacterium | NA | NA | 3316667 | 1.56 | 0.71 | 0.08 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | NA | 33166634 | 1.26 | 0.84 | 0.10 |
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Ruminococcaceae UCG − 005 | 33166612 | 0.88 | 0.62 | 0.07 |
| Verrucomicrobia | WCHB1–41 | uncultured bacterium | NA | NA | 33166611 | 0.62 | 0.30 | 0.04 |
| Firmicutes | Clostridia | Clostridiales | Family XIII | Mogibacterium | 33166640 | 0.61 | 0.32 | 0.04 |
aNA indicates that the taxonomic rank could not be annotated
bPercentage relative abundances are given in terms of the mean (n = 70), standard deviation (SD) and standard error of the mean (SEM)
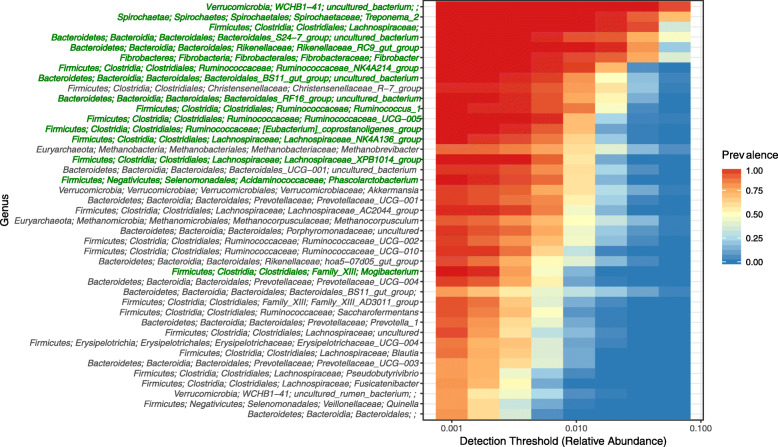
Similar analysis performed with the OTUs grouped at the genus level showed that 41 genus level groups were present in at least 75% of the animals when a cut-off of > 0.001 was used (Fig. 7). Of these 41 genus level groups, only 16 were core (Fig. 7; Table 2). Of the core genus level groups only five could be annotated to characterized genera: Treponema 2, Fibrobacter, Ruminococcus 1, Phascolarctobacterium and Mogibacterium. In contrast, little is known about the other 11 genus level groups which were represented by uncharacterized taxa at the genus (nine groups), family (one group) and class (one group) level. The two most predominant core genus level groups, consistent with the OTU based analysis, were an uncharacterized class belonging to the uncultivated class WCHB1–41 within the Verrucomicrobia and Treponema 2 (Table 2). These 16 core genus level groups represented 61.2% ± 8.35 (mean ± SD) of the prokaryotic community.
Fig. 7.
Heat map showing the relative abundance (> 0.001 cut-off) and prevalence (75% cut-off) of the prokaryotic genus level OTU groups in the 70 equine fecal samples analyzed. Different detection thresholds are used, providing information regarding the relative abundance of the genus level OTU groups relative to their prevalence. Taxonomic assignments of the genera are given to five taxonomic ranks (phylum, class, order, family and genus) where possible. Where this was not possible, the non-annotated ranks were left empty (e.g. Verrucomicrobia; WCHB1–41; uncultured_bacterium;;; has no information for the family and genus ranks). Genus level groups present in all animals have their taxonomic assignments written in green.
Table 2.
Taxonomic annotationa and percentage relative abundancesb of the core taxa detected at the genus level
| Phylum | Class | Order | Family | Genus | Mean | SD | SEM |
|---|---|---|---|---|---|---|---|
| Verrucomicrobia | WCHB1–41 | uncultured bacterium | NA | NA | 12.79 | 4.36 | 0.52 |
| Spirochaetae | Spirochaetes | Spirochaetales | Spirochaetaceae | Treponema 2 | 8.35 | 3.81 | 0.46 |
| Bacteroidetes | Bacteroidia | Bacteroidales | Bacteroidales S24–7 group | uncultured bacterium | 6.93 | 4.51 | 0.54 |
| Fibrobacteres | Fibrobacteria | Fibrobacterales | Fibrobacteraceae | Fibrobacter | 6.33 | 4.13 | 0.49 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | NA | 5.73 | 1.97 | 0.23 |
| Bacteroidetes | Bacteroidia | Bacteroidales | Rikenellaceae | Rikenellaceae RC9 gut group | 4.96 | 1.88 | 0.22 |
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Ruminococcaceae NK4A214 group | 2.35 | 0.94 | 0.11 |
| Bacteroidetes | Bacteroidia | Bacteroidales | Bacteroidales BS11 gut group | uncultured bacterium | 2.32 | 1.57 | 0.19 |
| Bacteroidetes | Bacteroidia | Bacteroidales | Bacteroidales RF16 group | uncultured bacterium | 2.01 | 1.14 | 0.14 |
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Ruminococcus 1 | 1.88 | 0.86 | 0.10 |
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Ruminococcaceae UCG-005 | 1.58 | 0.94 | 0.11 |
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | [Eubacterium] coprostanoligenes group | 1.36 | 0.60 | 0.07 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Lachnospiraceae NK4A136 group | 1.36 | 0.73 | 0.09 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Lachnospiraceae XPB1014 group | 1.29 | 0.66 | 0.08 |
| Firmicutes | Negativicutes | Selenomonadales | Acidaminococcaceae | Phascolarctobacterium | 1.29 | 0.66 | 0.08 |
| Firmicutes | Clostridia | Clostridiales | Family XIII | Mogibacterium | 0.65 | 0.36 | 0.04 |
aNA indicates that the taxonomic rank could not be annotated
bPercentage relative abundances are given in terms of the mean (n = 70), standard deviation (SD) and standard error of the mean (SEM)
No methanogenic archaeal taxon was present in all equines, neither at OTU- nor genus level. Of the seven archaeal genera, Methanocorpusculum (91.4%) and Methanobrevibacter (88.6%) were most prevalent in the animals sampled (Fig. 7), and represented (mean ± SD) 44.7 ± 30.83% and 51.2 ± 30.41% of the archaeal 16S rRNA gene sequences detected per animal, respectively. The large variation in the mean relative abundance was mainly due to the predominance of one of these two genera compared to the other, or in some animals only Methanocorpusculum (6 animals) or Methanobrevibacter (5 animals) being present. The other archaeal genera were all lower in prevalence (< 22.9%), and all represented a much smaller proportion (< 0.98%) of the archaeal 16S rRNA gene sequences detected per animal.
Similarly, no core anaerobic fungal OTUs or genera were observed across all equines. Of the eight genera Caecomyces (95.3%) and ALI (53.1%) were most predominant in terms of prevalence in the animals sampled, and represented (mean ± SD) 48.2 ± 39.26% and 31.6 ± 39.11% of the anaerobic fungal sequences detected per animal, respectively. The large variation in the mean relative abundance was due to animals being often predominated by only one of these two anaerobic fungal genera (Additional File 7: Figure S6). The other anaerobic fungal genera were all lower in prevalence (< 18.8%) and overall represented a much smaller proportion (< 5.62%) of the mean anaerobic fungal sequences detected per animal.
Discussion
The objectives of this study were (i) to give an overview of the fecal microbiota across different equine types in terms of bacteria, archaea and anaerobic fungi, and (ii) to determine core taxa. All of the equines studied contained archaea and anaerobic fungi as well as bacteria in their fecal microbiota. However, equine type was associated with differences in both microbial concentrations and community composition.
With respect to microbial concentrations, the largest difference observed between the equine types was in terms of the anaerobic fungi. The six-fold higher anaerobic fungal concentrations in donkey compared to horse is consistent with the reported higher fiber digestion ability of donkeys compared to horses [17]. As anaerobic fungi can take from 8 to 32 h to complete their life cycle [30], the longer fiber mean retention time in donkeys relative to that in horses is also likely to be more favorable for anaerobic fungal growth [17]. The anaerobic fungal concentration for the horse × donkey was numerically intermediate between donkey and horse. Whether this is due to horse × donkey having an intermediate fiber mean retention time compared to that of horses and donkeys is not known.
Archaeal concentrations in donkey and horse × donkey were both approximately two-fold higher compared to those in horse. As methanogenic archaea have been shown to increase the rate and extent of cellulose degradation by anaerobic fungi [31], this may result in enhanced anaerobic fungal activity and fiber degradation in donkey and horse × donkey. The archaeal and bacterial concentrations followed the same trend with equine type suggesting that the equine hindgut ecosystem may select for a certain bacteria:archaea ratio. The microbial concentrations in zebra did not significantly differ from those in horse. This is interesting as in genetic terms horse, zebras and asses represent three distinct lineages [15], and suggests that the gut physiology of zebra may be more comparable to that of horse than to that of donkey.
In general, donkey was the most distinctive among the equine types studied here. Associated with this, several bacterial genus level taxa were identified that were positively associated with donkey or the other three equine types, respectively. However, as the diet, age and management of the animals was not controlled, these initial observations regarding equine type require further investigation with more specifically designed studies. Nevertheless, the variation encompassed in the equine cohort studied here is valuable when determining the core microbiota of equine hindgut.
As with human studies [13], the number of bacterial OTUs reported to be core in the equine hindgut differs in the literature, and is influenced by a variety of factors including the number of individuals sampled as well as the approaches used to define operational taxonomic units (OTUs) and the core. The first study to report on the core bacteria in fecal samples found 123 OTUs common to four horses [3], whereas later more in depth studies reported smaller cores of 25 OTUs with ten horses/ponies [8], six OTUs with 17 horses [10], eight OTUs with 11 horses [9], 65 OTUs with 12 horses/ponies [11] and 21 OTUs with 35 ponies [7]. In this study, eight prokaryotic core OTUs were found to be shared in fecal samples from 70 different equines. These eight core OTUs represented a total of 13.4% of the prokaryotic community per animal. This is a much larger proportion of the fecal microbiota compared to the 2.3% [10] and 6.7% [7] of total sequences previously reported. This is perhaps not surprising considering that at least half of the core OTUs in this study belonged to phyla that were not detected in the core OTUs identified in the other studies.
A core OTU belonging to the phylum Spirochaetes was only found in the Morrison et al. [7] study, whereas no core OTU belonging to the Verrucomicrobia was found in any other study to date [3, 7, 8, 10, 11]. In fact, in these studies the phylum Verrucomicrobia was not detected at all despite it being reported in previous studies to represent 3% [32], 4.1% [2] and 18.1% [1] of the fecal microbiota of healthy horses. This phylum has also been reported to occur throughout the equine hindgut [9]. The reason for the earlier failure to detect Verrucomicrobia is not entirely clear, but has been previously suggested to be caused by methodological differences in the 16S rRNA gene regions and/or sequencing platforms used [9].
Following the characterization of the first cultured representative of the Verrucomicrobia subdivision 5 [33], this subdivision has now been re-classified as a new phylum named Kiritimatiellaeota [34]. One of the eight distinct phylogenetic clades within this phylum, RFP12, is comprised mainly of sequences retrieved from the intestine of vertebrates, e.g. the rumen of cattle [34] as well as equine feces [1]. Whilst the three published isolates from the phylum do not belong to the RFP12 clade, it is interesting to note that Kiritimatiella glycovorans has a strictly anaerobic and fermentative type of metabolism with sugars as preferred substrate [34]. The other two isolates can utilize sulfated polysaccharides including the glycoprotein mucin [35]. Manual re-annotation of the four Verrucomicrobia core OTUs with the latest version of the SILVA database (version 132) [36], confirmed that they do indeed belong to the Kiritimatiellaeota and are placed in a class called WCHB1–41. This class is named after a sequenced bacterial clone from the methanogenic zone of a contaminated aquifer [37]. In light of the sequences of Steelman et al. [1] being assigned to the phylogenetic clade RFP12 [34], it is speculated that the four Kiritimatiellaeota core OTUs identified in this study also belong to this RFP12 clade.
As these four core OTUs accounted for 7.2% of the mean fecal prokaryotic community/animal in this study, there is an urgent need to obtain cultured representatives of this clade in order to determine their role in the health and nutrition of mammalian herbivores. This is of particular interest, as it has been previously reported that Verrucomicrobia subdivision 5 was of higher relative abundance in horses suffering from laminitis compared to healthy controls [1], and that its relative abundance was dramatically decreased by the oral administration of the antibiotic trimethoprim-sulfadiazine [4].
The core OTU belonging to the Spirochaetes annotated as Treponema 2 accounted for 3.4% of the prokaryotic community/animal in this study, and at the genus level Treponema 2 comprised 8.4%. The genus Treponema was reported to be lower in relative abundance in other studies (1.9% [2] and 2.9% [1]) but consistent with this study was present in all 18 of the animals in the study of Steelman et al. [1]. In the study of Daly et al. [32], the majority of the Spirochaetes cloned sequences clustered with the known species Treponema bryantii and Treponema succinifaciens. Neither of these species are considered to be pathogenic, unlike several of the other 20 species listed within the Treponema 2 genus in the SILVA database [36]. Treponema bryantii utilizes fermentable substrates, in particular soluble sugars released from cellulose by cellulolytic bacteria such as Fibrobacter (formerly Bacteroides) succinogenes [38]. Treponema succinifaciens is strictly saccharolytic, can ferment pentoses, hexoses and disaccharides, and produces relatively large amounts of succinate from glucose and CO2 [39].
Of the three remaining core OTUs, only one could be annotated to a genus that has cultivated representatives: Mogibacterium. Considering that the five species belonging to this genus are all associated with oral disease [40, 41], its role in the equine hindgut ecosystem is unclear. This genus has been previously reported to occur in equine feces in some studies [2, 5, 7], but not in others [1]. This is perhaps not surprising considering this core taxon had the lowest relative abundance at both the OTU and genus level. The other two core OTUs were annotated as an unknown genus in the Lachnospiraceae and the uncultured genus Ruminococcaceae UCG − 005. Previous studies have consistently shown that bacterial OTUs belonging to the families Lachnospiraceae and Ruminococcaceae are normal members of the bacterial core of the equine hindgut [8, 10].
Only when the OTUs were grouped at the genus level were the well-known cellulolytic bacterial genera Fibrobacter and Ruminococcus 1 (containing R. albus and R. flavefaciens) found to be part of the core. Whilst it has been previously concluded that R. flavefaciens is more predominant in the equine hindgut than F. succinogenes [19], this was not indicated in the current study. The mean relative abundance of Fibrobacter was three times higher than that of Ruminococcus 1. As no single OTU belonging to these two genera was core, niche differentiation is likely to be important for the occurrence of particular OTUs. For example, in the study of Dougal et al. [10], core OTUs belonging to the family Fibrobacteraceae were found in animals fed a hay diet, but not with the other diets studied.
Two other bacterial genera with cultured representatives were identified as being core: Phascolarctobacterium and the [Eubacterium] coprostanoligenes group. As Eubacterium coprostanoligenes is a cholesterol reducing anaerobe [42], its role in the equine hindgut remains to be elucidated. The two species belonging to the genus Phascolarctobacterium have both been isolated from mammalian feces and utilize succinate, suggesting that this genus might also be involved in the metabolism of succinate in the equine hindgut [43, 44]. Other core genus level groupings of OTUs identified in this study belonged to uncharacterized genera in the families Lachnospiraceae, Rikenellaceae and Ruminococcaceae, as well as uncharacterized families in the order Bacteroidales. It is clear that cultured representatives of these taxa need to be isolated before it can be speculated what their role in the equine hindgut may represent.
Despite the numerous core bacterial OTUs and genera detected in this study, no core archaeal OTU or genus was observed. However, consistent with other studies the genera Methanocorpusculum and Methanobrevibacter were found to predominate [6, 27]. Both genera consist of species that mainly utilize carbon dioxide and hydrogen for the production of methane [45, 46], suggesting that functional redundancy may result in no archaeal taxon being core. This has been previously observed in ruminants where Methanobrevibacter is most prevalent, whereas Methanocorpusculum is not normally found in the rumen [47].
Similar to the archaea, no core anaerobic fungal OTU or genus was detected in this study. Findings of this study indicated that equine type was associated with differences in anaerobic fungal community composition and concentration. Caecomyces was predominant in the donkey where the highest anaerobic fungal concentrations were detected, whereas uncultivated AL1 predominated in the horse and zebra where the lowest anaerobic fungal concentrations were found. Further work is needed to confirm this finding with animals that are fed and managed in the same way.
The limited prevalence of Piromyces in the equine cohort studied here is in contrast to the fact that it has been the main genus identified in cultivation based studies to date [22, 48–51]. Caecomyces [52] and Buwchfawromyces [53] have also been cultured from equines. A culture independent survey indicated that two uncultivated taxa, currently termed AL1 and AL3 [29], were predominant in the eight equines (representing five different species) sampled [25]. A preliminary study has also shown that AL1 can be found throughout the equine hindgut [54]. As the majority of OTUs in this study could not be classified at the clade level, it is clear that novel anaerobic fungal taxa remain to be isolated from the equine hindgut.
Fecal samples are commonly used as a marker for the equine hindgut microbiota, as they can be obtained non-invasively. In terms of bacterial community composition analysis, fecal samples are generally similar to the different anatomical sections of the hindgut at the phylum and class levels [8, 9]. Indeed, fecal bacterial communities have been reported to not be significantly different from those in the colon [9], although they represent the distal regions of the hindgut to a more limited extent [8, 9, 28]. Preliminary studies have also reported differences in archaeal and anaerobic fungal community composition along the equine hindgut [54, 55]. As such, interpretation of the findings of this study relative to the proximal hindgut should be made cautiously.
Diet is also well known to influence the equine hindgut microbiome [56], and it has been previously reported that diet can influence the core microbiota [10]. Dougal et al. [10] found that animals fed a hay based diet that was either supplemented with a starch rich supplement or an oil supplement had a smaller core microbiota compared to animals fed an unsupplemented hay diet, and that only a limited core spanned all three diets. As Dougal et al. [10] did not detect any Verrucomicrobia, it is perhaps not surprising that the core present in all three of the diets provided in their study was represented by only 6 OTUs that accounted for 2.3% of total sequences. Furthermore, one Spirochaete OTU was only core in the oil supplemented hay based diet and not in the starch supplemented hay based diet or unsupplemented hay diet. It is clear from these contrasting findings, relative to the present study, that further work is needed to determine how the core microbiota in the hindgut of healthy equines is affected by diet.
Conclusions
Equine studies to date have primarily focused on one equine species and only the bacterial component of the hindgut. In this study, observations indicated that equine type (horse, donkey, horse × donkey and zebra) was associated with differences in both fecal microbial concentrations and community composition, with donkey generally being most distinctive. Despite this, a common bacterial core was found in all the equines studied that was larger than previously reported. This was primarily due to the detection of predominant core taxa belonging to the phyla Kiritimatiellaeota (formerly Verrucomicrobia subdivision 5) and Spirochaetes. Archaea and anaerobic fungi were present in all animals, although no core OTU or genus was detected for either. The lack of microbial cultures representing the core and predominant bacterial and anaerobic fungal taxa, respectively, needs to be addressed, particularly as both are likely to play a key role in the ability of equines to utilize dietary fiber. There is, therefore, an urgent need to culture and characterize representative key taxa to advance fundamental understanding of the microbial taxa that underpin the equine hindgut ecosystem.
Methods
Animals
Fecal samples were collected from 70 different equines that were grouped into four different equine types: horse (Equus ferus caballus), donkey (Equus africanus asinus), horse × donkey (Equus ferus caballus × Equus africanus asinus) and zebra (Equus zebra hartmannae, Equus quagga burchellii, Equus quagga boehmi and Equus grevyi). The horse × donkey animals sampled could not be classed as mules or hinnies due to lack of information about their parentage. For each equine type a representative animal size range was used, with the exception of zebras where this was not possible with the exception of variation between (sub)species. All animals sampled were 4–26 years old and had been clinically healthy in the 6 months prior to sampling, with no known history of gut-related problems. Animals for each equine type were sourced from multiple locations and belonged to either The Donkey Sanctuary, Utrecht University, private owners or zoos. The majority of the animals sampled had a predominantly pasture based diet at the time of sampling, with the exception of two horses and five of the zebras, which had no access to fresh pasture. Further animal specific details are provided in Additional file 1: (Table S1).
Fecal sample collection and determination of dry matter content
For each animal one freshly voided fecal sample was collected during either September or October 2016. Parts of the feces that were visibly free of dirt, bedding etc. were collected into a clean bucket, and then a pre-weighed tube was filled (approx. 20–30 g wet weight). The filled tubes were then weighed before being placed on wet ice. Samples were kept on wet ice for a maximum of one hour before being stored at − 20 °C. Fecal samples were then freeze-dried to a constant weight. For each sample, the percentage fecal dry matter content was then calculated using the original wet weight and the final freeze-dried weight.
DNA extraction
The freeze dried fecal material was broken up by hand, and any large fibrous particles cut into smaller pieces using a sterile scalpel. The material was then placed into a mortar and manually ground with a pestle. Total DNA was extracted from 25 mg of freeze-dried and ground fecal sample using a protocol involving a combination of bead beating, Stool Transport and Recovery (STAR) buffer (Roche Diagnostics Nederland BV, Almere, Netherlands) and the Maxwell® 16 Instrument (Promega, Leiden, Netherlands). The method was as previously published [57] except that the sample was first treated for 60 s at a speed of 6.0 m/s before adding the STAR buffer, to ensure all the sample material was finely ground. The purity of the resulting DNA extract was assessed using a NanoDrop ND-1000 spectrophotometer (NanoDrop® Technologies, Wilmington, DE, USA), and the quantity assessed using a Qubit dsDNA BR assay (Thermoscientific, Bleiswijk, Netherlands).
Determination of microbial concentrations
For absolute quantification of bacteria and archaea, SYBR green qPCR assays were performed with sample DNA extracts using a CFX384 Touch™ Real-Time PCR Detection System (Bio-Rad Laboratories BV, Veenendaal, Netherlands) as previously described [57]. All qPCR analyses were carried out in triplicate with a reaction volume of 10 μL. Bacterial and archaeal assays used 0.2 ng and 2 ng, respectively, of sample DNA. Equine specific standard curves (108 to 102 amplicon copies/μL) for the assays were prepared using purified PCR amplicons generated from an equine fecal DNA extract using the primers and cycling conditions previously described for the preparation of standards [57].
For absolute quantification of anaerobic fungi, a Taqman probe based method was used as previously described [58] with the exception that a CFX384 Touch™ Real-Time PCR Detection System (Bio-Rad Laboratories BV) was used. All qPCR analyses were carried out in triplicate with a reaction volume of 10 μL, and 2 ng of sample DNA was used. Standard curves (108 to 101 amplicon copies/μL) for the assays were prepared using purified PCR amplicons generated from Neocallimastix frontalis strain RE1 DNA (kindly provided by Dr. Tony Callaghan, Bavarian State Research Center for Agriculture, Freising, Germany). The PCR amplicon was generated using the primers Neo18SF (5′-AATCCTTCGGATTGGCT-3′: [58]) and AF LSU reverse (5′-CTTGTTAAMYRAAAAGTGCATT-3′: [59]).
Prokaryotic community composition analysis
For 16S rRNA gene based prokaryotic composition profiling, barcoded amplicons from the V4 region of 16S rRNA genes were generated from the DNA extracts using a 2-step PCR strategy as previously described [57]. Barcoded PCR products were mixed in equimolar amounts into pools together with defined synthetic mock communities which allow assessment of potential technical biases [60]. Pools were then sequenced on the Illumina HiSeq platform using the HiSeq Rapid Run 300 bp paired end (PE) sequencing mode (GATC-Biotech, Konstanz, Germany, now part of Eurofins Genomics Germany GmbH).
The 16S rRNA gene sequencing data was analyzed using NG-Tax [60]. NG-Tax defines OTUs using an open reference approach, and OTUs are defined as unique sequences that are above a user-defined minimum abundance threshold. NG-Tax (version NG-Tax-1.jar, which is available at http://download.systemsbiology.nl/ngtax/) was run with the following default settings: 70 nt read length, ratio OTU abundance 2.0, classify ratio 0.8, minimum percentage threshold 0.1%, identity level 100% and error correction of 1 mismatch (98.5%). Paired-end libraries were filtered to contain only read pairs with perfectly matching barcodes, and those barcodes were used to demultiplex reads by sample. Taxonomy was assigned to OTUs using the 128 version of the SILVA 16S rRNA gene reference database [36].
Anaerobic fungal composition analysis
For anaerobic fungal community composition profiling, barcoded amplicons comprising the partial 18S rRNA gene (~ 130 bp), full ITS1 region and partial 5.8S rRNA gene (~ 31 bp) were generated using a 2-step PCR strategy with a SensoQuest Labcycler (Göttingen, Germany) [61]. The first PCR step was performed with previously published ARISA primers [58] with the addition of UniTag adapters (underlined): Neo 18S For 5′-GAGCCGTAGCCAGTCTGCAATCCTTCGGATTGGCT-3′ and Neo 5.8S Rev. 5′-GCCGTGACCGTGACATCGCGAGAACCAAGAGATCCA-3′. PCR was performed in a total volume of 25 μL containing 1× HF buffer (Finnzymes, Vantaa, Finland), 1 μL dNTP Mix (10 mM; Promega), 1 U of Phusion® Hot Start II High-Fidelity DNA polymerase (Finnzymes), 500 nM of each primer and 2 ng of sample DNA. The cycling conditions consisted of an initial denaturation at 98 °C for 3 min followed by 40 cycles of 98 °C for 10 s, 58 °C for 30 s and 72 °C for 30 s, and then a final extension at 72 °C for 6 min. Triplicate PCR reactions were prepared for each sample, along with a non-template control (NTC) reaction. The presence of the PCR products was assessed by agarose gel electrophoresis on a 2% (w/v) agarose gel containing 1× SYBR® Safe (Invitrogen, Carlsbad, CA, USA). Pooled triplicate reactions, as well as the negative NTC reaction, were then purified using HighPrep™ (MagBio Europe Ltd., Kent, United Kingdom).
The second PCR step was then employed to add an 8 nucleotide sample specific barcode to the 5′- and 3′- end of the PCR products as previously described [57]. Each PCR reaction, with a final volume of 100 μL, contained 5 μL of the purified first step PCR product, 5 μL each of barcoded forward and reverse primers (10 μM), 2 μL dNTP Mix (10 mM), 2 U of Phusion® Hot Start II High-Fidelity DNA polymerase and 1× HF buffer. Amplification consisted of an initial denaturation at 98 °C for 30 s followed by 5 cycles of 98 °C for 10 s, 52 °C for 20 s and 72 °C for 20 s, and then a final extension at 72 °C for 10 min. Barcoded PCR products were then purified using the HighPrep™ and quantified using a Qubit in combination with the dsDNA BR Assay Kit (Invitrogen). Purified barcoded PCR products were then pooled in equimolar amounts along with defined synthetic mock communities [61]. Pools were then sequenced on the Illumina HiSeq platform using the HiSeq Rapid Run 300 bp PE sequencing mode (GATC-Biotech, Konstanz, Germany, now part of Eurofins Genomics Germany GmbH).
The anaerobic fungal sequence data was then analyzed using NG-Tax [61]. NG-Tax (version NGTax-2.jar which is available at http://download.systemsbiology.nl/ngtax/) was run using the default parameters (as described earlier) except for the following: 150 nt read length, minimum percentage threshold 0.6% and error correction of 1 mismatch (99.3%). As the barcoded amplicon primers used were not within the AF-ITS1 database used for OTU annotation (which is a requirement for annotation by NG-Tax), an empty database file (emptydb.fasta.gz which is available at http://download.systemsbiology.nl/ngtax/databases/) was used and the OTUs then subsequently annotated manually.
Fasta files of the OTUs from the NG-Tax generated biom file were extracted using the script OTUseq_export.py. The OTUs were annotated using BLASTN searches against the AF-ITS1 database [29] (version 3.3, available from www.anaerobicfungi.org) using default settings with “-num_alignments 10” (BLAST version 2.4.0). For OTUs that could not be annotated by the AF-ITS1 database, BLASTN searches were performed against the NCBI database. Cut-off levels for OTU annotations were determined based on the mean percentage similarities of full-length sequences in the AF-ITS1 database within clade and within genus. These cut-off levels were > 98% for clade and > 95% for genus. The NG-Tax generated biom file was converted to a tab delimited table to enable OTU annotations to be added. The OTUs that were clearly associated with the NTC sample were also manually removed from the tab delimited table at this stage, along with any OTUs that were not anaerobic fungal in origin. The resulting tab delimited table was then converted back to a biom file.
Statistical analysis
Microbial composition summary box plots, alpha diversity, UniFrac based Principal Coordinate Analysis (PCoA) and core microbiome analysis was performed within R (version 3.4.1) [62] using the following packages: microbiome (http://microbiome.github.com/microbiome), microbiomeutilities (https://github.com/microsud/microbiomeutilities), RColorBrewer [63], bindrcpp (https://github.com/krlmlr/bindrcpp), magrittr [64], phyloseq [65], picante [66], nlme [67], vegan [68], lattice [69], permute [70], ape [71], ggplot2 [72] and ggpubr [73]. The figures from ggplot2 and ggpubr were further refined in Adobe Illustrator CC (version 22.1) to improve their clarity. Redundancy analysis (RDA) was performed using Canoco 5 [74] to assess the relationship between genus-level phylogenetic groupings of the OTUs and equine type. Univariate data (fecal dry matter, alpha diversity and qPCR data) were analysed using a one-way ANOVA with equine type as a single independent factor and using a Tukey post-hoc test (Genstat 18th Edition, VSN International Ltd). Fecal dry matter and alpha diversity data was normally distributed, whereas the qPCR data was not normally distributed and, therefore, was analysed after Log10 transformation.
Supplementary information
Additional file 1: Table S1. Details of the animals used in the study.
Additional file 2: Figure S1. Effect of equine type on fecal dry matter content. Columns represent the mean (n = 18, except for zebra where n = 16) and error bars the SEM. Letters indicate significant differences (P < 0.05).
Additional file 3: Figure S2. Effect of equine type on the fecal bacterial (A), anaerobic fungal (B) and archaeal (C) concentrations on a fresh weight basis. Columns represent the mean (n = 18, except for zebra where n = 16) and error bars the SEM. Letters above the bars within each plot indicate significant differences (P < 0.05). Percentages stated on the x-axis in brackets indicate how the mean of each equine type compares to that of the horse.
Additional file 4: Figure S3. Boxplot showing the six main bacterial and archaeal phyla detected in the different equine types (minor phyla grouped as ‘Other’).
Additional file 5: Figure S4. Boxplot showing the twenty bacterial and archaeal families detected in the different equine types that were of highest relative abundance. Families are grouped by phylum as follows: (1) Bacteroidetes, (2) Euryarchaeota, (3) Fibrobacteres, (4) Firmicutes, (5) Spirochaetae and (6) Verrucomicrobia. Families that could not be classified are grouped as ‘Unclassified Families’.
Additional file 6: Figure S5. Boxplot showing the anaerobic fungal clades detected in the different equine types. Clades that could not be classified are grouped as ‘Unclassified Clades’.
Additional file 7: Figure S6. Boxplot showing the anaerobic fungal genera detected in the different equine types. Genera that could not be classified are grouped as ‘Unclassified Genera’.
Acknowledgements
The owners of the equines sampled are thanked for their participation in the study. Gerben Hermes and Edoardo Saccenti are also thanked for their constructive advice with respect to data analysis.
Abbreviations
- AF-ITS1
Anaerobic fungal ITS1
- ITS1
Internal Transcribed Spacer 1
- NTC
Non-template control
- OTU
Operational Taxonomic Unit
- PCoA
Principal co-ordinate analysis
- PCR
Polymerase chain reaction
- PE
Paired end
- qPCR
Quantitative PCR
- RDA
Redundancy analysis
- SD
Standard deviation
- SEM
Standard error of the mean
Author contributions
JE obtained funding and designed the study with support from HS, FB, DD, WP and JD. JE, PB and FB collected the samples. JE and PB performed the lab work. JE and SS performed the data analysis. All authors contributed to the writing of the manuscript and its revision. All authors read and approved the final manuscript.
Funding
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement number 706899.
Availability of data and materials
The datasets and material supporting the conclusions of this article are provided as follows. Additional information is provided in Additional files 1, 2, 3, 4, 5, 6 and 7. The barcoded amplicon sequence data is deposited in the European Nucleotide Archive under the study accession number PRJEB31377. All the sample barcodes, R codes, data used in the analysis and the workflow (as Rmarkdown files to reproduce the microbial community analysis) are available at https://github.com/mibwurrepo/EdwardsJ_2019_EquineCoreMicrobiome.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
J. E. Edwards, Email: joanee2002@hotmail.com
S. A. Shetty, Email: sudarshan.shetty@wur.nl
P. van den Berg, Email: paul.vandenberg@wur.nl
F. Burden, Email: faith.burden@thedonkeysanctuary.org.uk
D. A. van Doorn, Email: D.A.vanDoorn1@uu.nl
W. F. Pellikaan, Email: wilbert.pellikaan@wur.nl
J. Dijkstra, Email: jan.dijkstra@wur.nl
H. Smidt, Email: hauke.smidt@wur.nl
Supplementary information
Supplementary information accompanies this paper at 10.1186/s42523-020-0023-1.
References
- 1.Steelman SM, Chowdhary BP, Dowd S, Suchodolski J, Janečka JE. Pyrosequencing of 16S rRNA genes in fecal samples reveals high diversity of hindgut microflora in horses and potential links to chronic laminitis. BMC Vet Res. 2012;8:231. doi: 10.1186/1746-6148-8-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shepherd ML, Swecker WS, Jensen RV, Ponder MA. Characterization of the fecal bacteria communities of forage-fed horses by pyrosequencing of 16S rRNA V4 gene amplicons. FEMS Microbiol Lett. 2012;326:62–68. doi: 10.1111/j.1574-6968.2011.02434.x. [DOI] [PubMed] [Google Scholar]
- 3.Costa MC, Arroyo LG, Allen-Vercoe E, Stämpfli HR, Kim PT, Sturgeon A, et al. Comparison of the fecal microbiota of healthy horses and horses with colitis by high throughput sequencing of the V3-V5 region of the 16S rRNA gene. PLoS One. 2012;7:e41484. doi: 10.1371/journal.pone.0041484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costa MC, Stämpfli HR, Arroyo LG, Allen-Vercoe E, Gomes RG, Weese J. Changes in the equine fecal microbiota associated with the use of systemic antimicrobial drugs. BMC Vet Res. 2015;11:19. doi: 10.1186/s12917-015-0335-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodriguez C, Taminiau B, Brévers B, Avesani V, Van Broeck J, Leroux A, et al. Faecal microbiota characterisation of horses using 16 rdna barcoded pyrosequencing, and carriage rate of Clostridium difficile at hospital admission. BMC Microbiol. 2015;15:181. doi: 10.1186/s12866-015-0514-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernandes KA, Kittelmann S, Rogers CW, Gee EK, Bolwell CF, Bermingham EN, et al. Faecal microbiota of forage-fed horses in New Zealand and the population dynamics of microbial communities following dietary change. PLoS One. 2014;9:e112846. doi: 10.1371/journal.pone.0112846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morrison PK, Newbold CJ, Jones E, Worgan HJ, Grove-White DH, Dugdale AH, et al. The equine gastrointestinal microbiome: impacts of age and obesity. Front Microbiol. 2018;9:3017. doi: 10.3389/fmicb.2018.03017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dougal K, de la Fuente G, Harris PA, Girdwood SE, Pinloche E, Newbold CJ. Identification of a Core bacterial community within the large intestine of the horse. PLoS One. 2013;8:e77660. doi: 10.1371/journal.pone.0077660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costa MC, Silva G, Ramos RV, Staempfli HR, Arroyo LG, Kim P, et al. Characterization and comparison of the bacterial microbiota in different gastrointestinal tract compartments in horses. Vet J. 2015;205:74–80. doi: 10.1016/j.tvjl.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 10.Dougal K, de la Fuente G, Harris PA, Girdwood SE, Pinloche E, Geor RJ, et al. Characterisation of the Faecal bacterial Community in Adult and Elderly Horses fed a high fibre, high oil or high starch diet using 454 pyrosequencing. PLoS One. 2014;9:e87424. doi: 10.1371/journal.pone.0087424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dougal K, Harris PA, Girdwood SE, Creevey CJ, Curtis GC, Barfoot CF, et al. Changes in the total fecal bacterial population in individual horses maintained on a restricted diet over 6 weeks. Front Microbiol. 2017;8:1502. doi: 10.3389/fmicb.2017.01502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blackmore TM, Dugdale A, Argo CM, Curtis G, Pinloche E, Harris PA, et al. Strong stability and host specific bacterial Community in Faeces of ponies. PLoS One. 2013;8:e75079. doi: 10.1371/journal.pone.0075079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shetty SA, Hugenholtz F, Lahti L, Smidt H, de Vos WM. Intestinal microbiome landscaping: insight in community assemblage and implications for microbial modulation strategies. FEMS Microbiol Rev. 2017;41:182–199. doi: 10.1093/femsre/fuw045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banerjee S, Schlaeppi K, van der Heijden MGA. Keystone taxa as drivers of microbiome structure and functioning. Nat Rev Microbiol. 2018;16:567–576. doi: 10.1038/s41579-018-0024-1. [DOI] [PubMed] [Google Scholar]
- 15.Jónsson H, Schubert M, Seguin-Orlando A, Ginolhac A, Petersen L, Fumagalli M, et al. Speciation with gene flow in equids despite extensive chromosomal plasticity. PNAS. 2014;111:18655–18660. doi: 10.1073/pnas.1412627111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.FAOSTAT. 2016. http://faostat.fao.org/. Accessed 23 Nov 2016.
- 17.Cuddeford D, Pearson RA, Archibald RF, Muirhead R. Digestibility and gastro-intestinal transit time of diets containing different proportions of alfalfa and oat straw given to thoroughbreds, Shetland ponies, Highland ponies and donkeys. Anim Sci. 1995;61:407–417. doi: 10.1017/S1357729800013953. [DOI] [Google Scholar]
- 18.Smith DG, Pearson RA. A review of the factors affecting the survival of donkeys in semi-arid regions of sub-Saharan Africa. Trop Anim Health Prod. 2005;37:1–19. doi: 10.1007/s11250-005-9002-5. [DOI] [PubMed] [Google Scholar]
- 19.Julliand V, Grimm P. Horse species symposium: the microbiome of the horse hindgut: history and current knowledge. J Anim Sci. 2016;94:2262–2274. doi: 10.2527/jas.2015-0198. [DOI] [PubMed] [Google Scholar]
- 20.Lee SS, Ha JK, Cheng KJ. Relative contributions of bacteria, protozoa, and fungi to in vitro degradation of orchard grass cell walls and their interactions. Appl Environ Microbiol. 2000;66:3807–3813. doi: 10.1128/AEM.66.9.3807-3813.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gruninger RJ, Puniya AK, Callaghan TM, Edwards JE, Youssef N, Dagar SS, et al. Anaerobic fungi (phylum Neocallimastigomycota): advances in understanding their taxonomy, life cycle, ecology, role and biotechnological potential. FEMS Microbiol Ecol. 2014;90:1–17. doi: 10.1111/1574-6941.12383. [DOI] [PubMed] [Google Scholar]
- 22.Solomon KV, Haitjema CH, Henske JK, Gilmore SP, Borges-Rivera D, Lipzen A, et al. Early-branching gut fungi possess large, comprehensive array of biomass-degrading enzymes. Science. 2016;351:1192–1195. doi: 10.1126/science.aad1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng YF, Edwards JE, Allison GG, Zhu WY, Theodorou MK. Diversity and activity of enriched ruminal cultures of anaerobic fungi and methanogens grown together on lignocellulose in consecutive batch culture. Bioresour Technol. 2009;100:4821–4828. doi: 10.1016/j.biortech.2009.04.031. [DOI] [PubMed] [Google Scholar]
- 24.Moore BE, Dehority BA. Effects of diet and hindgut defaunation on diet digestibility and microbial concentrations in the cecum and colon of the horse. J Anim Sci. 1993;71:3350–3358. doi: 10.2527/1993.71123350x. [DOI] [PubMed] [Google Scholar]
- 25.Liggenstoffer AS, Youssef NH, Couger MB, Elshahed MS. Phylogenetic diversity and community structure of anaerobic gut fungi (phylum Neocallimastigomycota) in ruminant and non-ruminant herbivores. ISME J. 2010;4:1225–1235. doi: 10.1038/ismej.2010.49. [DOI] [PubMed] [Google Scholar]
- 26.Julliand V, Riondet C, De Vaux A, Alcaraz G, Fonty G. Comparison of metabolic activities between Piromyces citronii, an equine fungal species, and Piromyces communis, a ruminal species. Anim Feed Sci Technol. 1998;70:161–168. doi: 10.1016/S0377-8401(97)00043-6. [DOI] [Google Scholar]
- 27.Lwin K-O, Matsui H. Comparative analysis of the methanogen diversity in horse and pony by using mcrA gene and Archaeal 16S rRNA gene clone libraries. Archaea. 2014;2014:483574. doi: 10.1155/2014/483574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dougal K, Harris PA, Edwards A, Pachebat JA, Blackmore TM, Worgan HJ, et al. A comparison of the microbiome and the metabolome of different regions of the equine hindgut. FEMS Microbiol Ecol. 2012;82:642–652. doi: 10.1111/j.1574-6941.2012.01441.x. [DOI] [PubMed] [Google Scholar]
- 29.Koetschan C, Kittelmann S, Lu J, Al-Halbouni D, Jarvis GN, Müller T, et al. Internal transcribed spacer 1 secondary structure analysis reveals a common core throughout the anaerobic fungi (Neocallimastigomycota) PLoS One. 2014;9:e91928. doi: 10.1371/journal.pone.0091928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gordon GLR, Phillips MW. The role of anaerobic gut fungi in ruminants. Nutr Res Rev. 1998;11:133–168. doi: 10.1079/NRR19980009. [DOI] [PubMed] [Google Scholar]
- 31.Mountfort DO, Asher RA, Bauchop T. Fermentation of cellulose to methane and carbon dioxide by a rumen anaerobic fungus in a triculture with Methanobrevibacter sp. strain RA1 and Methanosarcina barkeri. Appl Environ Microbiol. 1982;44:128–134. doi: 10.1128/AEM.44.1.128-134.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daly K, Stewart CS, Flint HJ, Shirazi-Beechey SP. Bacterial diversity within the equine large intestine as revealed by molecular analysis of cloned 16S rRNA genes. FEMS Microbiol Ecol. 2001;38:141–151. doi: 10.1111/j.1574-6941.2001.tb00892.x. [DOI] [Google Scholar]
- 33.Hugenholtz P, Goebel BM, Pace NR. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/JB.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spring S, Bunk B, Spröer C, Schumann P, Rohde M, Tindall BJ, et al. Characterization of the first cultured representative of Verrucomicrobia subdivision 5 indicates the proposal of a novel phylum. ISME J. 2016;10:2801–2816. doi: 10.1038/ismej.2016.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Vliet DM, Ayudthaya PN, Diop S, Villanueva S, Stams L, Sánchez-Andrea I A. Anaerobic degradation of sulfated polysaccharides by two novel Kiritimatiellales strains isolated from Black Sea sediment. Front Microbiol. 2019;10:253. doi: 10.3389/fmicb.2019.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dojka MA, Hugenholtz P, Haack SK, Pace NR. Microbial diversity in a hydrocarbon- and chlorinated-solvent- contaminated aquifer undergoing intrinsic bioremediation. Appl Environ Microbiol. 1998;64:3869–3877. doi: 10.1128/AEM.64.10.3869-3877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stanton TB, Canale-Parola E. Treponema bryantii sp. nov., a rumen spirochete that interacts with cellulolytic bacteria. Arch Microbiol. 1980;127:145–156. doi: 10.1007/BF00428018. [DOI] [PubMed] [Google Scholar]
- 39.Cwyk WM, Canale-Parola E. Treponema succinifaciens sp. nov., an anaerobic spirochete from the swine intestine. Arch Microbiol. 1979;122:231–239. doi: 10.1007/BF00411285. [DOI] [PubMed] [Google Scholar]
- 40.Nakazawa F, Poco SE, Sato M, Ikeda T, Kalfas S, Sundqvist G, et al. Taxonomic characterization of Mogibacterium diversum sp. nov. and Mogibacterium neglectum sp. nov., isolated from human oral cavities. Int J Syst Evol Microbiol. 2002;52:115–122. doi: 10.1099/00207713-52-1-115. [DOI] [PubMed] [Google Scholar]
- 41.Nakazawa F, Sato M, Poco SE, Hashimura T, Ikeda T, Kalfas S, et al. Description of Mogibacterium pumilum gen. Nov., sp. nov. and Mogibacterium vescum gen. Nov., sp. nov., and reclassification of Eubacterium timidum (Holdeman et al. 1980) as Mogibacterium timidum gen. Nov., comb. nov. Int J Syst Evol Microbiol. 2000;50:679–688. doi: 10.1099/00207713-50-2-679. [DOI] [PubMed] [Google Scholar]
- 42.Freier TA, Beitz DC, Li L, Hartman PA. Characterization of Eubacterium coprostanoligenes sp. nov., a cholesterol-reducing anaerobe. Int J Syst Bacteriol. 2009;44:137–142. doi: 10.1099/00207713-44-1-137. [DOI] [PubMed] [Google Scholar]
- 43.Del Dot T, Osawa R, Stackebrandt E. Phascolarctobacterium faecium gen. Nov, spec. Nov., a novel taxon of the Sporomusa group of Bacteria. Syst Appl Microbiol. 1993;16:380–384. doi: 10.1016/S0723-2020(11)80269-9. [DOI] [Google Scholar]
- 44.Watanabe Y, Nagai F, Morotomi M. Characterization of Phascolarctobacterium succinatutens sp. Nov., an asaccharolytic, succinate-utilizing bacterium isolated from human feces. Appl Environ Microbiol. 2012;78:511–518. doi: 10.1128/AEM.06035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zellner G, Stackebrandt E, Messner P, Tindall BJ, Conway de Macario E, Kneifel H, et al. Methanocorpusculaceae fam. nov., represented by Methanocorpusculum parvum, Methanocorpusculum sinense spec. nov. and Methanocorpusculum bavaricum spec. nov. Arch Microbiol. 1989;151:381–390. doi: 10.1007/BF00416595. [DOI] [PubMed] [Google Scholar]
- 46.Leadbetter JR, Breznak JA. Physiological ecology of Methanobrevibacter cuticularis sp. nov. and Methanobrevibacter curvatus sp. nov., isolated from the hindgut of the termite Reticulitermes flavipes. Appl Environ Microbiol. 1996;62:3620–3631. doi: 10.1128/AEM.62.10.3620-3631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Henderson G, Cox F, Ganesh S, Jonker A, Young W, Janssen PH, et al. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci Rep. 2015;5:14567. doi: 10.1038/srep14567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Orpin CG. Isolation of cellulolytic Phycomycete Fungi from the Caecum of the horse. Microbiology. 1981;123:287–296. doi: 10.1099/00221287-123-2-287. [DOI] [PubMed] [Google Scholar]
- 49.Freelove ACJ, Bolam DN, White P, Hazlewood GP, Gilbert HJ. A novel carbohydrate-binding protein is a component of the plant Cell Wall-degrading complex of Piromyces equi. J Biol Chem. 2001;276:43100–43117. doi: 10.1074/jbc.M107143200. [DOI] [PubMed] [Google Scholar]
- 50.Gaillard-Martinie B, Breton A, Dusser M, Julliand V. Piromyces citronii sp. nov., a strictly anaerobic fungus from the equine caecum: a morphological, metabolic, and ultrastructural study. FEMS Microbiol Lett. 1995;130:321–326. doi: 10.1111/j.1574-6968.1995.tb07738.x. [DOI] [PubMed] [Google Scholar]
- 51.Li J, Heath IB, Bauchop T. Piromyces mae and Piromyces dumbonica, two new species of uniflagellate anaerobic chytridiomycete fungi from the hindgut of the horse and elephant. Can J Bot. 1990;68:1021–1033. doi: 10.1139/b90-129. [DOI] [Google Scholar]
- 52.Gold JJ, Brent Heath I, Bauchop T. Ultrastructural description of a new chytrid genus of caecum anaerobe, Caecomyces equi gen. nov., sp. nov., assigned to the Neocallimasticaceae. BioSystems. 1988;21:403–415. doi: 10.1016/0303-2647(88)90039-1. [DOI] [PubMed] [Google Scholar]
- 53.Callaghan TM, Podmirseg SM, Hohlweck D, Edwards JE, Puniya AK, Dagar SS, et al. Buwchfawromyces eastonii gen. nov., sp. nov.: a new anaerobic fungus (Neocallimastigomycota) isolated from buffalo faeces. MycoKeys. 2015;9:11–28. doi: 10.3897/mycokeys.9.9032. [DOI] [Google Scholar]
- 54.Mura E, Edwards J, Kittelmann S, Kaerger K, Voigt K, Mrázek J, et al. Anaerobic fungal communities differ along the horse digestive tract. Fungal Biol. 2019;123:240–246. doi: 10.1016/j.funbio.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 55.Fliegerova K, Mura E, Mrázek J, Moniello G. A comparison of microbial profiles of different regions of the equine hindgut. Livest Sci. 2016;190:18–19. doi: 10.1016/j.livsci.2016.05.015. [DOI] [Google Scholar]
- 56.Julliand V, Grimm P. The impact of diet on the hindgut microbiome. J Equine Vet Sci. 2017;52:23–28. doi: 10.1016/j.jevs.2017.03.002. [DOI] [Google Scholar]
- 57.van Lingen HJ, Edwards JE, Vaidya JD, van Gastelen S, Saccenti E, van den Bogert B, et al. Diurnal dynamics of gaseous and dissolved metabolites and microbiota composition in the bovine rumen. Front Microbiol. 2017;8:425. doi: 10.3389/fmicb.2017.00425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Edwards JJE, Kingston-Smith AH, Jimenez HR, Huws SA, Skøt KP, Griffith GW, et al. Dynamics of initial colonization of nonconserved perennial ryegrass by anaerobic fungi in the bovine rumen. FEMS Microbiol Ecol. 2008;66:537–545. doi: 10.1111/j.1574-6941.2008.00563.x. [DOI] [PubMed] [Google Scholar]
- 59.Dollhofer V, Callaghan TM, Griffith GW, Lebuhn M, Bauer J. Presence and transcriptional activity of anaerobic fungi in agricultural biogas plants. Bioresour Technol. 2017;235:131–139. doi: 10.1016/j.biortech.2017.03.116. [DOI] [PubMed] [Google Scholar]
- 60.Ramiro-Garcia J, Hermes GDA, Giatsis C, Sipkema D, Zoetendal EG, Schaap PJ, et al. NG-Tax, a highly accurate and validated pipeline for analysis of 16S rRNA amplicons from complex biomes. F1000Research. 2016;5:1791. doi: 10.12688/f1000research.9227.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edwards, Joan E.; Hermes, GDA; Kittelmann, S; Nijsse, B.; Smidt H. Assessment of the Accuracy of Anaerobic Fungal ITS1 Based Barcoded Amplicon Sequencing for Community Composition Analysis. Submitted. [DOI] [PMC free article] [PubMed]
- 62.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2013. URL http://www.R-project.org/
- 63.Neuwirth E. RColorBrewer: ColorBrewer Palettes. R package version 1.1–2. 2014. https://cran.r-project.org/package=RColorBrewer
- 64.Milton Bache S, Wickham H. magrittr: A Forward-Pipe Operator for R. R package version 1.5. 2014. https://cran.r-project.org/package=magrittr
- 65.McMurdie PJ, Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, et al. Picante: R tools for integrating phylogenies and ecology. Bioinformatics. 2010;26:1463–1464. doi: 10.1093/bioinformatics/btq166. [DOI] [PubMed] [Google Scholar]
- 67.Pinheiro J, Bates D, DebRoy S, Sarkar D, R Core Team. _nlme: Linear and Nonlinear Mixed Effects Models_. R package version 3.1–131. 2017. https://cran.r-project.org/package=nlme
- 68.Oksanen J, Guillaume Blanchet F, Friendly M, Kindt R, Legendre P, McGlinn D, et al. vegan: Community Ecology Package. R package version 2.5–3. 2018. https://cran.r-project.org/package=vegan
- 69.Deepayan S. Lattice: multivariate data visualization with R. New York: Springer; 2008. [Google Scholar]
- 70.Simpson GL. permute: Functions for Generating Restricted Permutations of Data. 2016. https://cran.r-project.org/package=permute
- 71.Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 72.Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer-Verlag; 2016. [Google Scholar]
- 73.Kassambara A. ggpubr: “ggplot2” Based Publication Ready Plots. R package version 0.2. https://cran.r-project.org/package=ggpubr
- 74.Šmilauer, P., and Leps J. Multivariate analysis of ecological data using Canoco 5. Second Edition. New York: Cambridge University Press; 2014.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Details of the animals used in the study.
Additional file 2: Figure S1. Effect of equine type on fecal dry matter content. Columns represent the mean (n = 18, except for zebra where n = 16) and error bars the SEM. Letters indicate significant differences (P < 0.05).
Additional file 3: Figure S2. Effect of equine type on the fecal bacterial (A), anaerobic fungal (B) and archaeal (C) concentrations on a fresh weight basis. Columns represent the mean (n = 18, except for zebra where n = 16) and error bars the SEM. Letters above the bars within each plot indicate significant differences (P < 0.05). Percentages stated on the x-axis in brackets indicate how the mean of each equine type compares to that of the horse.
Additional file 4: Figure S3. Boxplot showing the six main bacterial and archaeal phyla detected in the different equine types (minor phyla grouped as ‘Other’).
Additional file 5: Figure S4. Boxplot showing the twenty bacterial and archaeal families detected in the different equine types that were of highest relative abundance. Families are grouped by phylum as follows: (1) Bacteroidetes, (2) Euryarchaeota, (3) Fibrobacteres, (4) Firmicutes, (5) Spirochaetae and (6) Verrucomicrobia. Families that could not be classified are grouped as ‘Unclassified Families’.
Additional file 6: Figure S5. Boxplot showing the anaerobic fungal clades detected in the different equine types. Clades that could not be classified are grouped as ‘Unclassified Clades’.
Additional file 7: Figure S6. Boxplot showing the anaerobic fungal genera detected in the different equine types. Genera that could not be classified are grouped as ‘Unclassified Genera’.
Data Availability Statement
The datasets and material supporting the conclusions of this article are provided as follows. Additional information is provided in Additional files 1, 2, 3, 4, 5, 6 and 7. The barcoded amplicon sequence data is deposited in the European Nucleotide Archive under the study accession number PRJEB31377. All the sample barcodes, R codes, data used in the analysis and the workflow (as Rmarkdown files to reproduce the microbial community analysis) are available at https://github.com/mibwurrepo/EdwardsJ_2019_EquineCoreMicrobiome.