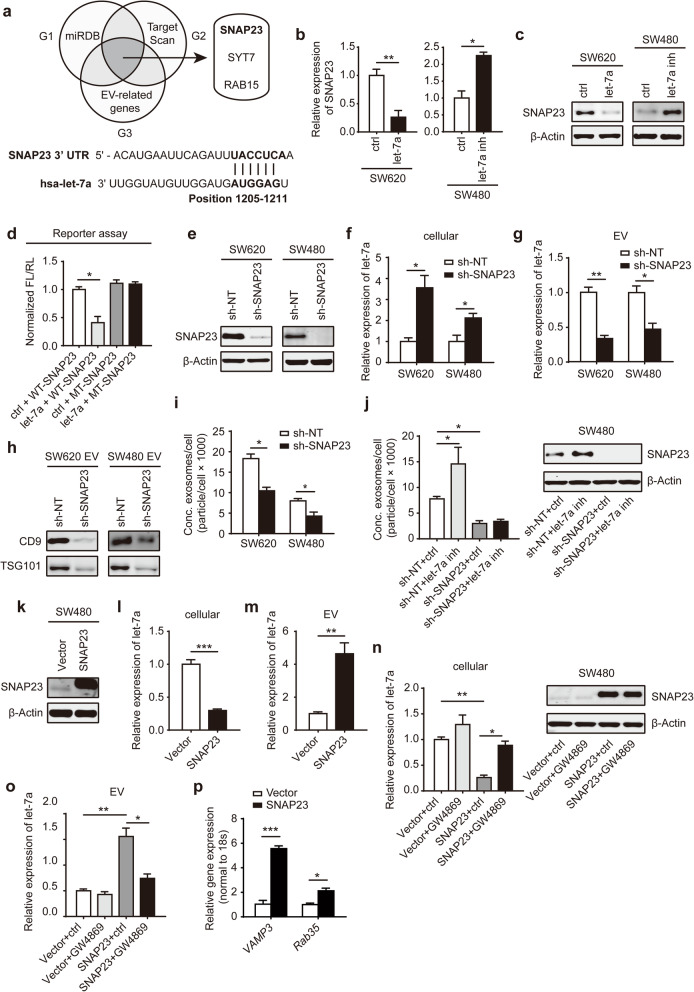
Fig. 3.
SNAP23 directly regulates the secretion of let-7a in EVs. a Venn diagram for selecting candidate target genes of let-7a using the three indicated data sets. G1, G2 predicted targets from miRDB (n = 990 genes) and Targetscan (n = 1207 genes); G3, EV secretion-related genes (n = 31). Summary of let-7a target sites in the 3’ UTRs of SNAP23 in the below. qPCR (b) and western blot analysis (c) of SNAP23 were determined in CRC cells expressing let-7a mimic or inhibitor, with U6 or β-Actin as the loading control. d Luciferase reporter assays confirmed the direct binding of let-7a to SNAP23. WT, wild type; MT, mutant. e Western blot analysis of SNAP23 in sh-NT and sh-SNAP23 CRC cells. f qPCR of intracellular let-7a from sh-NT and sh-SNAP23 CRC cells. g qPCR of extracellular let-7a in EVs from sh-NT and sh-SNAP23 CRC cells. h Western blot analysis of indicated proteins in EVs from CRC cells transfected with sh–NT and sh-SNAP23. i The ability of sh-SNAP23 CRC cells on EV secretion dramatically decreased compared to sh-NT CRC cells. j The effect on EV secretion (left) and SNAP23 expression (right) of SW480 sh-NT and sh-SNAP23 cells treated with ctrl or let-7a inhibitor. k Western blot analysis of SNAP23 in SW480 cells stably overexpressing empty vector and SNAP23. qPCR of intracellular let-7a (l) and extracellular let-7a (m) in SW480 cells overexpressing vector and SNAP23. n qPCR of intracellular let-7a (left) and western blotting of SNAP23 (right) from SW480 overexpressing vector and SNAP23 treated with vehicle ctrl or 10 μM GW4869. o qPCR of extracellular let-7a in EVs from SW480 overexpressing vector and SNAP23 treated with vehicle ctrl or GW4869. p qPCR analysis of related genes in SNARE complex from SW480 cells overexpressing vector and SNAP23. Data represent the mean ± SEM of at least three independent experiments. *p < 0.05, **p < 0.01 and ***p < 0.001