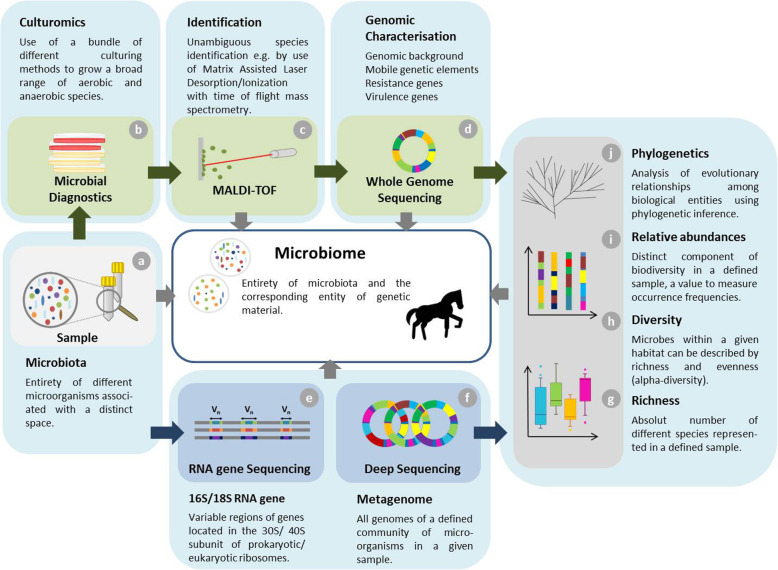
Fig. 1.
Workflow and synergistic application of differt techniques to study enteral microbiomes. Integrative and synergistic workflow to study equine microbiomes starting with dividing the fresh sample (a) for two general processing’s, microbial diagnostic in terms of culturomics (b-d) [157] and DNA sequencing approaches (e and f) for population analysis (g-i). A broad range of different aerob and aerobic culture conditions are used to initiate growth for microbial diagnostic (b), followed by rapid species identification by MALDI-tof mass spectrometry (c). Genome sequencing (d) allows (novel) species identification in case MALDI-tof provided no confident result or if resistance- and virulence encoding genes [158, 159] or other factors are of particular interest within a species. Both information sources allow identification of bacterial species present in the horse microbiota and their growth conditions. The second part of the sample should be stored native at − 80 °C until DNA extraction starts for either sequencing of variable regions of 16S/18S rRNA gene (e) allowing characterizing and quantifying taxonomic entities or sequencing of all genomes (metagenome) present in a sample (f). Further bioinformatics include description of richness (g), diversity indices (h) [160–162], relative abundances (i) and phylogenetics (j). Combination of classical diagnostics on a large scale and different techniques available to generate genomic data enable deep insights into microbiome composition and characteristics [163]