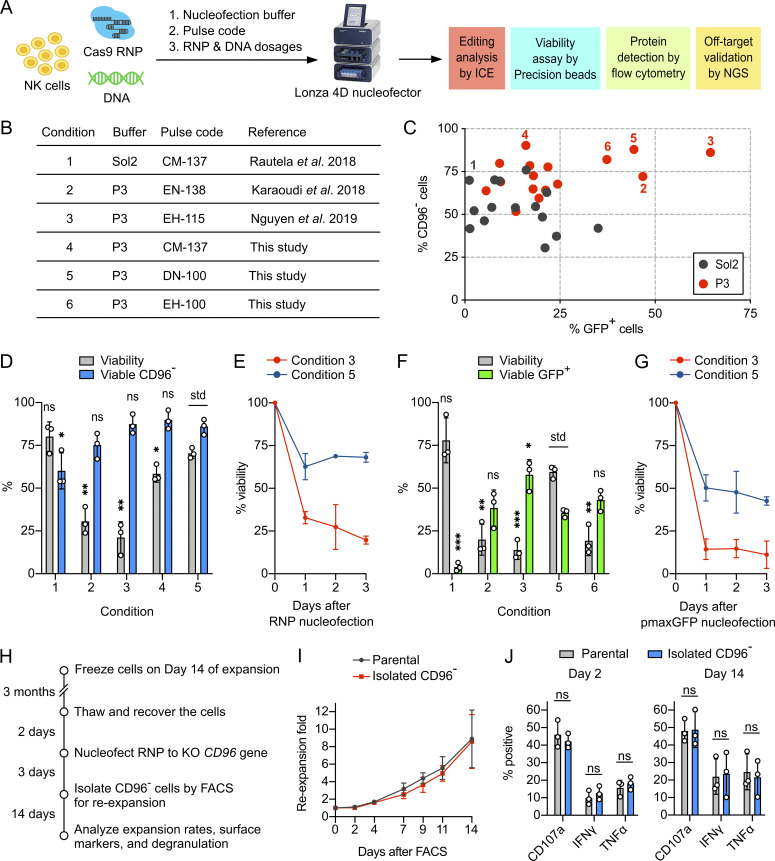
Figure 3.
Nucleofection optimization identified conditions for efficient and viable delivery of Cas9 RNP and DNA. (A) Workflow of the nucleofection screening and editing analyses to identify the optimal combination of nucleofection buffer, pulse code, and payload dosages. The screening was performed on NK cells from day 14 of the expansion. (B) Summary of the reference and newly identified conditions. (C) Screening for Cas9 RNP and DNA (using pmaxGFP reporter plasmid) nucleofection conditions in Sol2 and Lonza P3 buffer. CD96− and GFP+ cells were determined independently. The pulse codes, buffers, and raw data are listed in Table S1. Numerically labeled conditions are listed in the summary table. (D) Selected conditions were repeated for Cas9 RNP nucleofection to determine cell viability and viable CD96− cells by Precision beads assay. (E) Cell viability was monitored for 3 d after Cas9 RNP nucleofection. (F) Selected conditions were repeated for pmaxGFP nucleofection. (G) Cell viability after pmaxGFP nucleofection. (H) Workflow of the CD96 KO, FACS, and reexpansion. (I) Reexpansion rates of the FACS-isolated CD96− and parental cells. (J) Analysis of degranulation markers after chemical stimulation. Data are shown as mean ± SD of three donors (n = 3). Two-tailed Welch’s unequal variances t test was used to test for statistical significance. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. ns, not significant; std, standard for comparison.