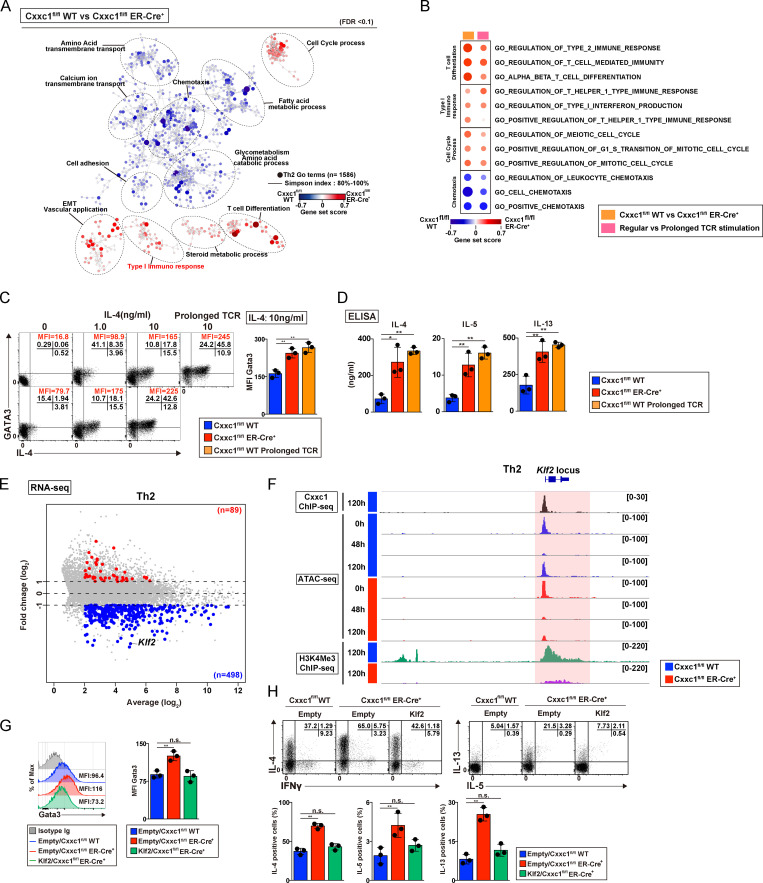
Figure 4.
Loss of Cxxc1 results in the enhanced expression of Th2 cytokines in CD4+ T cells. (A) The pathways that were changed in Cxxc1-deficient Th2 cells in comparison to WT control cells are shown. Dashed ellipses (added manually) indicate groups of similar GO terms. The node color indicates the gene set score. EMT, epithelial-to-mesenchymal transition. (B) The pathways changed in both Cxxc1-deficient Th2 cells and Th2 cells that received prolonged TCR/coreceptor stimulation are shown. (C) The representative profiles of IL-4 and GATA3 are shown. Naive CD4+ T cells from the indicated mice were cultured under Th2 cell–inducing conditions with the indicated concentrations of IL-4. The cultured cells were restimulated with PMA plus ionomycin for 4 h. A bar graph shows GATA3 mean fluorescence intensity (MFI) with SDs from three independent experiments (**, P < 0.01). (D) The protein expression of the indicated cytokines from WT Th2 cells, Cxxc1-deficient Th2 cells, or Th2 cells that received prolonged TCR/coreceptor stimulation. The indicated cytokines secreted by Th2 cells stimulated with anti-TCRβ for 24 h were measured by ELISA. Data from three independent experiments are shown with mean values and SDs (*, P < 0.05; **, P < 0.01). (E) The MA plot depicts 10,235 genes in WT versus Cxxc1-deficient Th2 cells based on RNA-seq. The blue and red points indicate Cxxc1-dependent group 4 genes that were down-regulated and up-regulated (greater than twofold) in the absence of Cxxc1, respectively. (F) A genome browser view of ChIP-seq signals in Th2 cells for 3xFlag-Cxxc1 and H3K4Me3 is shown for the Trib3 gene. Retroviral transduction of 3xFlag-Cxxc1 into Th2 cells was performed before ChIP-seq analysis. ATAC-seq signals of naive (0 h) and differentiation process (48 h and 120 h) of WT or Cxxc1-deficient cells are also shown. (G) Representative flow cytometry histograms of the GATA3 expression are shown (left). A bar graph shows GATA3 MFI with SDs from three independent experiments (**, P < 0.01; right). (H) Representative profiles of IFNγ versus IL-4 or IL-5 versus IL-13 in the indicated cells restimulated with PMA plus ionomycin for 4 h are shown (top). The percentages of IL-4–, IL-5–, and IL-13–positive CD4+ T cells are shown with the mean values and SDs (**, P < 0.01; bottom). n.s., not significant.