Figure 5. Loss of Chi3l1 alters microglial activation and enhances Aβ phagocytosis.
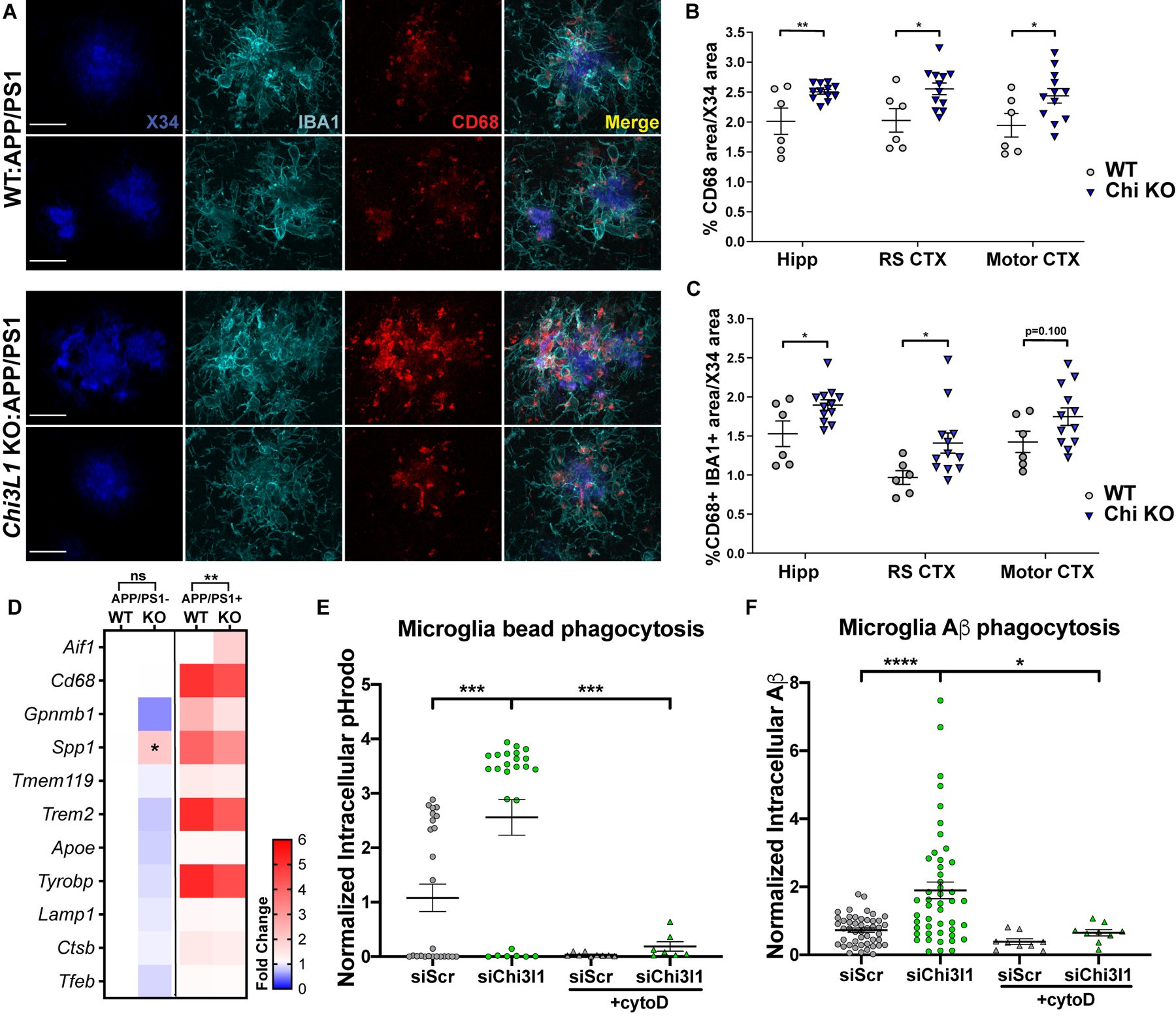
A. Representative high magnification images from hippocampi of 8 mo Chi3l1−/−:APP/PS1+ and APP/PS1+ control mice stained for X34 (fibrillar plaques), IBA1 (microglia), and CD68 (phagocytic microglia). Scale bar = 20μm
B–C. Quantification of CD68 (B) or colocalized IBA1/CD68 (C) from mice in (A) normalized to X34+ area in same section. Quantified from widefield image in Fig. S6. n = 6 (Chi3l1+/+) and 12 (Chi3l1−/−) mice per group. RS = retrosplenial
D. Microglia-associated gene expression from Fluidigm qPCR of 8 mo Chi3l1−/−:APP/PS1−, WT:APP/PS1−, Chi3l1−/−:APP/PS1+ and APP/PS1+ control mouse hippocampus. Mean of 4 mice (APP/PS1−) or 6–10 (APP/PS1+) mice per group normalized to WT:APP/PS1−. Two-way ANOVA with Tukey correction for multiple comparisons
E–F. pHrodo-labeled zymosan bead (E) or TAMRA-Aβ (F) uptake by primary microglia cultures transfected with control (siSCR) or Chi3l1 (siChi3l1) siRNA, +/− cytochalasin D to inhibit phagocytosis (+cytoD). Each point represents one field of view with an average of 209 (E) or 56 (F) cells/field. Data from 2 (E) or 1 (F) independent experiments.
All data represent mean +/− SEM. *p < 0.05, **p < 0.01, ***p < 0.001. Analyzed by two-tailed students t-test (B–C) or one-way ANOVA (E–F). All data subjected to Sidak correction for multiple comparisons unless otherwise noted.
