Figure 6.
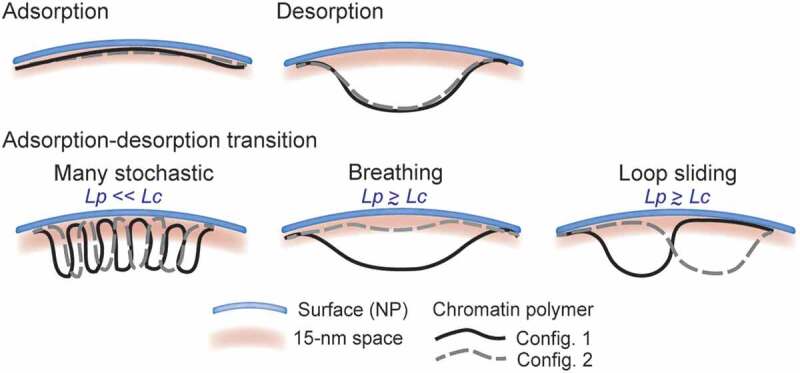
Interpretations of polymer simulations on chromatin configurations at the nuclear lamina. The genomic scale of our models (~50 kb) predicts chromatin behavior at the lamina within LADs rather than the behavior of entire LADs. We note however that simulations may also model narrow LADs of < 100 kb [11,14,15,36]. Monte Carlo simulations of the behavior of chromatin anchored at both ends to the lamina reveal various configurations depending on the ‘attraction strength’ of the system. A strong attraction potential induces a strong adsorption regime where the entire chromatin segment fully interacts with the lamina as a single ‘train’ (Adsorption). The absence of, or a very weak, attraction strength results in a desorbed chromatin segment which is only pined to the lamina by the preexisting anchors (Desorption). Within a LAD, such desorbed regions model a micro-loop domain of chromatin not bound to lamins, a configuration which has been as proposed earlier [21] and supported by the genomic and biochemical heterogeneity of LADs [20]. Between these adsorption and desorption extremes, weak but significant attraction potentials result in chromatin adsorption-desorption transition regimes. These can be interpreted as (i) many short-lived stochastic chromatin interactions with the lamina at any position along the segment (left); here, LP is greatly inferior to the chromatin contour length LC, which characterizes euchromatin. (ii) Lamina contacts and releases of an entire chromatin segment occur (except at the anchors), yielding a ‘breathing’ pattern (middle). (iii) Lamina contacts with and releases of sub-domains of relatively constant size but at varying positions occur along the segment, hence a ‘sliding loop’ pattern (right). The latter patterns are observed with LP approximating or greater than LC, modeling the behavior of a typical heterochromatic LAD sub-domain
