Figure 1.
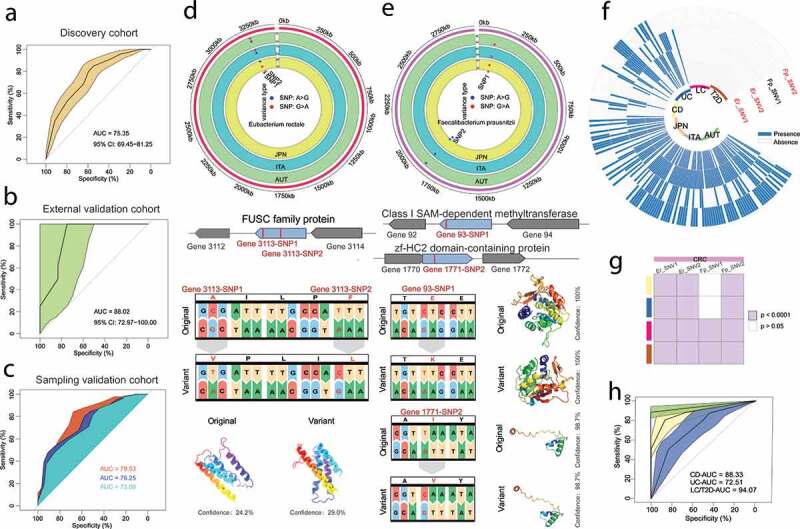
Construction and validation of CRC prediction model based on SNVs. (a) Prediction power in discovery cohorts with the accuracy of 75.35% using 22 SNV markers, including 4 SNVs enriched and 18 SNVs absent in CRC patients. (b) CRC classification accuracy for independent validation set recruited with 8 CRC patients and 12 healthy controls from Hainan Province, China. (c) The performance of the model in resampling validation cohorts. The sampling validation cohorts refer to the random sampling of 100 samples from discovery cohorts. (d) And (e) The related position, functions, amino acid and protein structure and of the four SNVs enriched in CRC. (f) And (g) Heatmaps were used to show the disease specificity of the four SNVs mentioned above, and three SNVs were enriched in CRC cohorts, including Er_SNV1, Er_SNV2 and FP_SNV2. (h) The accuracy of enriched SNVs to classify various diseases, and the accuracy ranged from 72.51% to 94.01%
