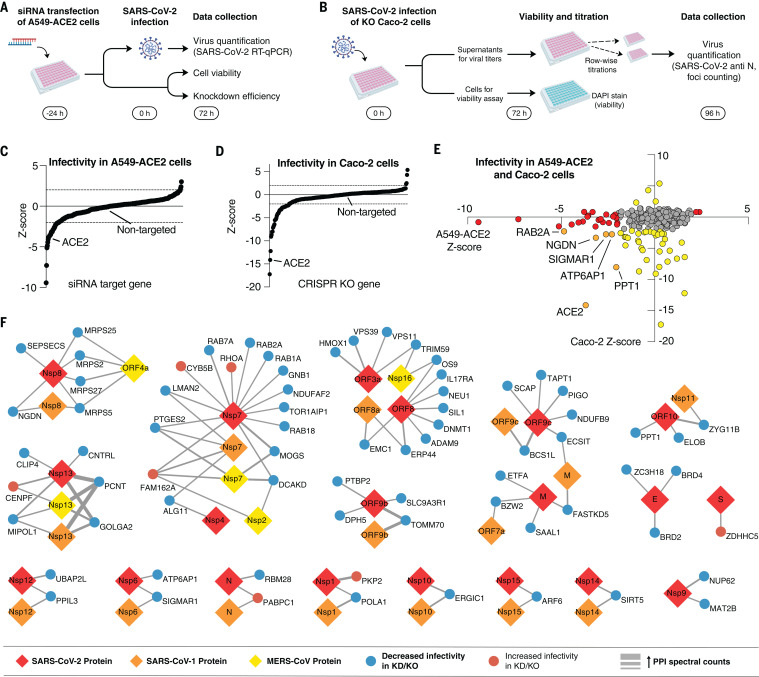
Fig. 5. Functional interrogation of SARS-CoV-2 interactors using genetic perturbations.
(A) A549-ACE2 cells were transfected with siRNA pools targeting each of the human genes from the SARS-CoV-2 interactome, followed by infection with SARS-CoV-2 and virus quantification using RT-qPCR. Cell viability and knockdown efficiency in uninfected cells was determined in parallel. (B) Caco-2 cells with CRISPR knockouts (KO) of each human gene from the SARS-CoV-2 interactome were infected with SARS-CoV-2, and supernatants were serially diluted and plated onto Vero E6 cells for quantification. Viabilities of the uninfected CRISPR knockout cells after infection were determined in parallel by DAPI staining. (C and D) Plot of results from the infectivity screens in A549-ACE2 knockdown cells (C) and Caco-2 knockout cells (D) sorted by z-score (z < 0, decreased infectivity; z > 0 increased infectivity). Negative controls (nontargeting control for siRNA, nontargeted cells for CRISPR) and positive controls (ACE2 knockdown or knockout) are highlighted. (E) Results from both assays with potential hits highlighted in red (A549-ACE2), yellow (Caco-2), and orange (both). (F) Pan-coronavirus interactome reduced to human preys with significant increase (red nodes) or decrease (blue nodes) in SARS-CoV2 replication upon knockdown or knockout. Viral proteins baits from SARS-CoV-2 (red), SARS-CoV-1 (orange), and MERS-CoV (yellow) are represented as diamonds. The thickness of the edge indicates the strength of the PPI in spectral counts. KD, knockdown; KO, knockout; PPI, protein-protein interaction.