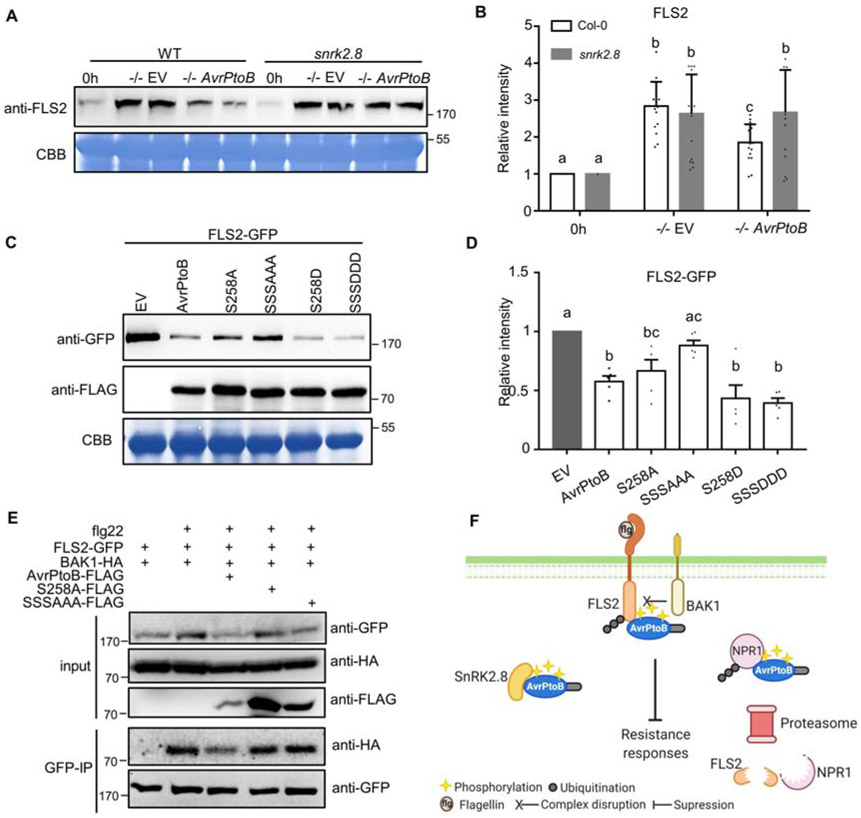
Figure 4. AvrPtoB-mediated degradation of FLS2 and inhibition of FLS2 complex formation requires the plant kinase SnrK2.8 and phosphorylated residues.
(A) FLS2 accumulation in Arabidopsis Col-0 and the snrk2.8 knockout after inoculation with P. syringae pv. tomato DC3000ΔavrPtoΔavrPtoB (−/−) variants. DC3000 −/− carrying the empty vector (EV) or a plasmid expressing wild-type AvrPtoB were syringe infiltrated into Col-0 and snrk2.8 at a concentration of 1 × 108 CFU mL−1 and proteins extracted after 8h. Protein extracts were subjected to anti-FLS2 immunoblotting. Coomassie Brilliant Blue (CBB) staining shows equal protein loading.
(B) Quantification of FLS2 band intensity in (A). FLS2 band intensities were quantified by Image Lab 6.0.1. The values were normalized first by Rubisco bands and subsequently by the intensities of “0h” bands. Data are means ± SD (n = 14 leaves). Different letters indicate significant differences (one-way ANOVA, Tukey’s test, p < 0.05).
(C) FLS2 accumulation in the presence of AvrPtoB phosphorylation mutants in N. benthamiana after Agrobacterium-mediated transient expression. 35S::FLS2-GFP was co-expressed with FLAG-tagged Dex-inducible AvrPtoB, AvrPtoBS258A, AvrPtoBS205AS210As258A (SSSAAA), AvrPtoBS258D, or AvrPtoBS205DS210DS258D (SSSDDD). The Dex inducible EV was used as a control. The expression of AvrPtoB-FLAG phosphorylation variants was induced by 15 μM DEX for 5 hours 24h post-Agrobacterium infiltration. Protein extracts were subjected to anti-GFP and anti-FLAG immunoblotting. CBB staining shows protein loading.
(D) Quantification of FLS2-GFP band intensity in (C). FLS2-GFP bands intensities were quantified by Image Lab 6.0.1. The values were normalized first by Rubisco bands and subsequently by the intensities of “EV” treatment. The data from two independent experiments were used for statistical analysis. Data are means ± SD (n = 6 leaves). Different letters indicate significant differences (one-way ANOVA, Tukey’s test, p < 0.05).
(E) Analyses of AvrPtoB-mediated inhibition of FLS2 complex formation. AvrPtoB-FLAG variants, BAK1-HA, and FLS2-GFP were co-expressed in the indicated combinations in N. benthamiana using Agrobacterium-mediated transient expression. The expression of AvrPtoB-FLAG phosphorylation variants was induced by 15 μM DEX for 3 hours 24h post-Agrobacterium infiltration. Leaves were infiltrated with 10 μM flg22 peptide 15 minutes before harvesting protein extracts for GFP immunoprecipitation and western blotting. Protein inputs and immunoprecipitated samples were detected by immunoblot.
(F) Model of AvrPtoB phosphorylation by SnRK2.8. Created with Biorender.com.